
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,114,619 – 23,114,767 |
| Length | 148 |
| Max. P | 0.901335 |

| Location | 23,114,619 – 23,114,727 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.14 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -22.46 |
| Energy contribution | -21.43 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23114619 108 + 27905053 UUUGU------GAUCGGCGGA-----CGCACAAGGAAUCAAUCAUGGGCCUGCUAUAUGCGCUGCGCGUGAGGAUCAUGAACUUCAUGAUUUUCUUCCUCCUCAUCAUCCUAAUGCCCG ...((------(((.((.(((-----(((.(((((..(((....))).)))((.....))..)).))))((((((((((.....))))))))))..)).))..)))))........... ( -28.70) >DroVir_CAF1 1562 115 + 1 UUAGU----GGAAAGGGCGGGUCCAACACAUACGUACGUAGCCAUGGGUCUGCUGAAGGCGUUGCGUGUGAGGAUCAUGAAUUUCAUGAUUUUCUUUCUAAUUAUCAUUUUGCUGCCGG .(((.----.(((..(((((..(((.....(((....)))....)))..)))))(((((((...)))..((((((((((.....)))))))))))))).........)))..))).... ( -33.20) >DroPse_CAF1 4990 114 + 1 UUUGUAUUUAGAAUCGGCAGA-----CGCACAACACACCAACCAUGGGCCUGCUUCGAGCCUUGCGUUUGAGAAUCAUGAAUUUCAUGGUUUUCUUCCUGAUAGUUAUAAUCCUGCCCA ...............((((((-----((((...............(((((......).)))))))))..((((((((((.....))))))))))..................))))).. ( -27.96) >DroEre_CAF1 1518 108 + 1 UUUGU------GAUCGGCGGA-----CGCACAAGAAAUCAAUCAUGGGCCUGUUGUACGCGCUGCGUGUGAGGAUCAUGAACUUCAUGAUUUUCUUCCUCAUCAUCAUCCUGAUGCCCG (((((------(..((.....-----))))))))...........((((......(((((...))))).((((((((((.....))))))))))......((((......)))))))). ( -28.00) >DroAna_CAF1 1590 108 + 1 CUUUA------GGCCGGUAGA-----UCCAAAACGAACUAAUCAUGGGUCUGCUGUAUGCUCUACGCGUGAGGAUCAUGAAUUUCAUGAUCUUCUUUCUGAUUAUCAUUCUGCUGCCCG .....------((((((((((-----((((..............))))))))))).((((.....))))((((((((((.....))))))))))....................))).. ( -34.04) >DroPer_CAF1 4990 114 + 1 UUUGUUGUUAGAAUCGGCAGA-----CGCACAACACACCAACCAUGGGCCUGCUUCGAGCCUUGCGUUUGAGAAUCAUGAACUUCAUGGUUUUCUUCCUAAUAGUUAUAAUCCUGCCCA ...............((((((-----((((...............(((((......).)))))))))..((((((((((.....))))))))))..................))))).. ( -27.96) >consensus UUUGU______AAUCGGCAGA_____CGCACAACAAACCAACCAUGGGCCUGCUGUAAGCGCUGCGUGUGAGGAUCAUGAACUUCAUGAUUUUCUUCCUAAUAAUCAUACUGCUGCCCG ...............(((((.......................(((.((..((.....))...)).)))((((((((((.....))))))))))..................))))).. (-22.46 = -21.43 + -1.02)



| Location | 23,114,647 – 23,114,767 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.89 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -20.40 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23114647 120 + 27905053 AAUCAUGGGCCUGCUAUAUGCGCUGCGCGUGAGGAUCAUGAACUUCAUGAUUUUCUUCCUCCUCAUCAUCCUAAUGCCCGGUCUGCCGCCGCGCACCACCUUCCCCUUCAAGGACUAUAU ....((((.(((((.....))(.((((((.((((((((((.....)))))))))).......................(((....))).)))))).).............))).)))).. ( -32.30) >DroVir_CAF1 1597 120 + 1 AGCCAUGGGUCUGCUGAAGGCGUUGCGUGUGAGGAUCAUGAAUUUCAUGAUUUUCUUUCUAAUUAUCAUUUUGCUGCCGGGUCUGCCCCCACGCACCACUUUCCCCUAUAAAGAUUUCAC ......(((......(((((.(.((((((.((((((((((.....)))))))))).......................(((....))).)))))).).)))))))).............. ( -32.80) >DroGri_CAF1 1573 120 + 1 AACAAUGGGCCUGCUCAAAGCGUUGCGUGUGCGAAUCAUGAAUUUCAUGAUAUUCUAUCUAAUUGUUAUUCUGUUGCCCGGUUUGCCACCGCGCACCACUUUCCCCUACAAGGAAUUCAC ......(((...((.....))..((.(((((((.((((((.....))))))............................((....))..))))))))).....))).............. ( -25.40) >DroMoj_CAF1 1494 120 + 1 AAUCAUGGGACUGUUGCGGUCAAUACGUCUGAGAAUUAUGAAUUUCAUGAUUUUCUUUCUAUUUAUUAUUUUGUUCCCGGGUCUGCCCCCGCGCACAACUUUCCCCUAUAAAGAUUUCAU ......((((.((((((((...........((((((((((.....)))))))))).......................(((....)))))))).)))....))))............... ( -27.50) >DroAna_CAF1 1618 120 + 1 AAUCAUGGGUCUGCUGUAUGCUCUACGCGUGAGGAUCAUGAAUUUCAUGAUCUUCUUUCUGAUUAUCAUUCUGCUGCCCGGUCUGCCUCCCCGUACCACCUUUCCCUUCAAGGAAUAUAU ......((((..((...((((.....))))((((((((((.....)))))))))).................)).))))(((.((......)).)))....((((......))))..... ( -30.10) >DroPer_CAF1 5024 120 + 1 AACCAUGGGCCUGCUUCGAGCCUUGCGUUUGAGAAUCAUGAACUUCAUGGUUUUCUUCCUAAUAGUUAUAAUCCUGCCCAACGUGCCGCCUCGCACCACUUUCCCCUACAAAGAAUUUAU .....(((((........(((..(......((((((((((.....))))))))))......)..)))........)))))..((((......))))........................ ( -24.99) >consensus AACCAUGGGCCUGCUGAAAGCGUUGCGUGUGAGAAUCAUGAAUUUCAUGAUUUUCUUUCUAAUUAUCAUUCUGCUGCCCGGUCUGCCCCCGCGCACCACUUUCCCCUACAAAGAAUUCAU ......(((...((.....))..((((((.((((((((((.....)))))))))).......................(((....))).))))))........))).............. (-20.40 = -21.23 + 0.84)

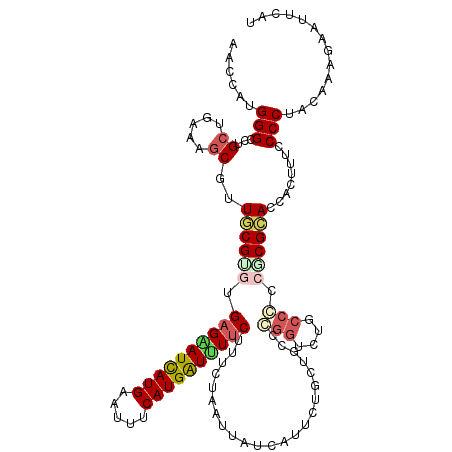
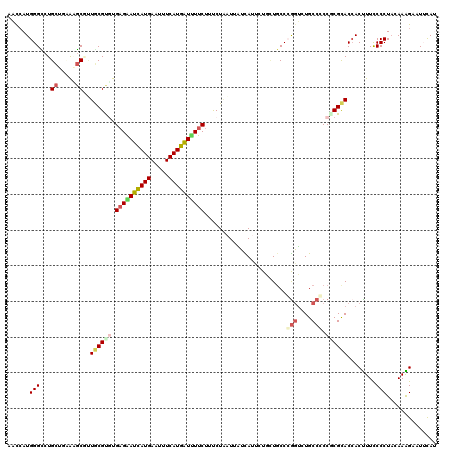
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:13 2006