
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,109,962 – 23,110,122 |
| Length | 160 |
| Max. P | 0.907011 |

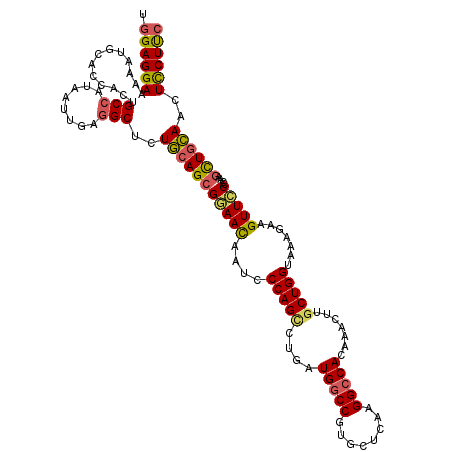
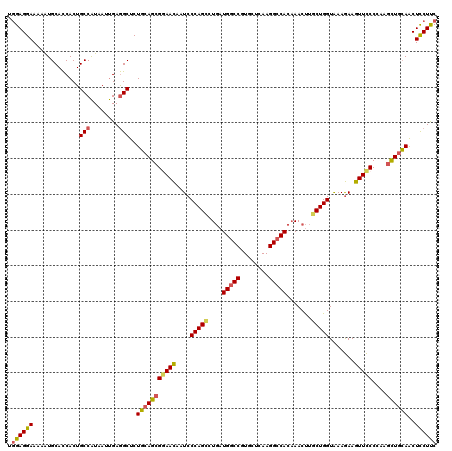
| Location | 23,109,962 – 23,110,082 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -31.10 |
| Energy contribution | -31.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23109962 120 + 27905053 UGGAGGAAAAAUGCACCACUGCCAUAAUUGAGGCUCUGCAGCGCAACAAUCCCAGCCUGAUGGCCGUGCUGAAGGCCACAAACUUGCUGGUAAAGAAGUUCCCCAAGCUGCAACUCCUUC .((((((.............(((........)))..((((((.........(((((.((.(((((........))))))).....)))))....(........)..))))))..)))))) ( -35.10) >DroSim_CAF1 15153 120 + 1 UGGAGGAAAAAUGCACCACUGCCAUAAUUGAGGCUCUGCAGCGGAACAAUCCCAGCCUGAUGGCCGUGCUCAAGGCCACAAACUUGCUGGUAAAGAAGUUCCCCAAGCUGCAACUCCUUC .((((((.............(((........)))..(((((((((((....(((((.((.(((((........))))))).....))))).......)))))....))))))..)))))) ( -40.20) >DroEre_CAF1 13479 120 + 1 UGGAGGAAAAAUGCACCACUGCCAUAAUUGAGGCUCUGCAGCGGAACAAUCCCAGCCUGAUGGCCGUGCUCAAGGCCACAAACUUGCUGGUAAAGAAGUUCCCCAAGCUGCAACUCCUUC .((((((.............(((........)))..(((((((((((....(((((.((.(((((........))))))).....))))).......)))))....))))))..)))))) ( -40.20) >DroYak_CAF1 13437 120 + 1 UGGAGGAAAAAUGCACCACUGCCAUAAUUGAGGCUCUGCAGCGGAACAAUCCCAGUCUGAUGGCCGUGCUCAAGGCCACAAACUUGCUGGUAAAGAAGUUCCCCAAGCUGCAACUCCUUC .((((((.............(((........)))..(((((((((((....(((((.((.(((((........))))))).....))))).......)))))....))))))..)))))) ( -38.60) >DroAna_CAF1 12864 120 + 1 UAGAGGAAAAGUGCAUCACUGCCGUUCUCGAAGCUCUCAAGAGAAAUAGUCCCAGUCUUUUGCCCAUUUUGAAGGCCACUAACCUUCUGGUGAAAAAGUUUCCGAGAUUGGAGCUUCUCC ..(((((..((((...))))....)))))((((((((....))).......((((((((..(((.........)))(((((......)))))...........)))))))).)))))... ( -31.20) >consensus UGGAGGAAAAAUGCACCACUGCCAUAAUUGAGGCUCUGCAGCGGAACAAUCCCAGCCUGAUGGCCGUGCUCAAGGCCACAAACUUGCUGGUAAAGAAGUUCCCCAAGCUGCAACUCCUUC .((((((.............(((........)))..(((((((((((....(((((....(((((........))))).......))))).......)))))....))))))..)))))) (-31.10 = -31.18 + 0.08)

| Location | 23,109,962 – 23,110,082 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -34.04 |
| Energy contribution | -35.00 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23109962 120 - 27905053 GAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUCAGCACGGCCAUCAGGCUGGGAUUGUUGCGCUGCAGAGCCUCAAUUAUGGCAGUGGUGCAUUUUUCCUCCA ...(((....(((((((..((((((((.....)).))))))((((((........)))))))))))))(((....(((((..(...(((........))).)..)))))....)))))). ( -45.20) >DroSim_CAF1 15153 120 - 1 GAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGGCUGGGAUUGUUCCGCUGCAGAGCCUCAAUUAUGGCAGUGGUGCAUUUUUCCUCCA ...(((((((((((....(((((.......(((((......((((((........))))))...)))))....)))))))))))).(((........)))...............)))). ( -44.80) >DroEre_CAF1 13479 120 - 1 GAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGGCUGGGAUUGUUCCGCUGCAGAGCCUCAAUUAUGGCAGUGGUGCAUUUUUCCUCCA ...(((((((((((....(((((.......(((((......((((((........))))))...)))))....)))))))))))).(((........)))...............)))). ( -44.80) >DroYak_CAF1 13437 120 - 1 GAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGACUGGGAUUGUUCCGCUGCAGAGCCUCAAUUAUGGCAGUGGUGCAUUUUUCCUCCA ...(((((((((((....(((((.......((....(((((((((((........))))).)))))).))...)))))))))))).(((........)))...............)))). ( -42.70) >DroAna_CAF1 12864 120 - 1 GGAGAAGCUCCAAUCUCGGAAACUUUUUCACCAGAAGGUUAGUGGCCUUCAAAAUGGGCAAAAGACUGGGACUAUUUCUCUUGAGAGCUUCGAGAACGGCAGUGAUGCACUUUUCCUCUA (((((((((((((((((((...(((((.(.(((((((((.....))))))....)))).))))).)))))).........))).)))))))(((((..(((....)))..))))).))). ( -37.00) >consensus GAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGGCUGGGAUUGUUCCGCUGCAGAGCCUCAAUUAUGGCAGUGGUGCAUUUUUCCUCCA ...(((((((((((....(((((.......(((((......((((((........))))))...)))))....)))))))))))).(((........)))...............)))). (-34.04 = -35.00 + 0.96)

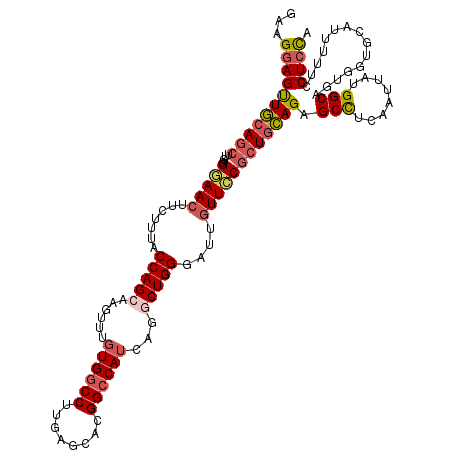
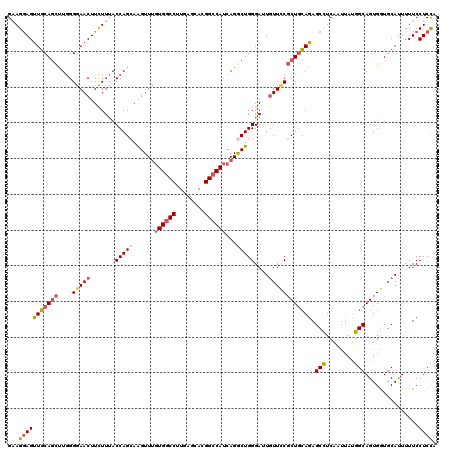
| Location | 23,110,002 – 23,110,122 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -47.36 |
| Consensus MFE | -34.54 |
| Energy contribution | -36.14 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23110002 120 - 27905053 UGGCGUUGCAGGCAGUGAUAGAAGUGCUCCAACUUCAGCGGAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUCAGCACGGCCAUCAGGCUGGGAUUGUUGCGC ..((((.((((((.((((.((((((.((((((((.((((.......)))).)).)))))).))))))))))..)).(((((((((((........))))).))))))....)))).)))) ( -46.40) >DroSim_CAF1 15193 120 - 1 UGGCGUUGCAGGCAAUGAUAGAAGUGCUCCAGCUUCAGCGGAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGGCUGGGAUUGUUCCGC ..(((..((((.(......((((((.((((((((.((((.......)))).))).))))).))))))...(((((......((((((........))))))...)))))).))))..))) ( -47.10) >DroEre_CAF1 13519 120 - 1 UGGCGUUGCAGGCAGUGAUAGAAGUGCUCCAGCUUCAGCGGAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGGCUGGGAUUGUUCCGC ..(((..((((((.((((.((((((.((((((((.((((.......)))).))).))))).))))))))))..)).(((((((((((........))))).))))))....))))..))) ( -48.00) >DroYak_CAF1 13477 120 - 1 UGGCGUUGCAGGCAGUGAUAGAAGUGCUCCAGCUUCAGCGGAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGACUGGGAUUGUUCCGC ..(((..((((((.((((.((((((.((((((((.((((.......)))).))).))))).))))))))))..)).(((((((((((........))))).))))))....))))..))) ( -48.50) >DroAna_CAF1 12904 120 - 1 UGCCGCUGCAGGCAGCGGUAGAAGUGCUCCAGGCGCAGGCGGAGAAGCUCCAAUCUCGGAAACUUUUUCACCAGAAGGUUAGUGGCCUUCAAAAUGGGCAAAAGACUGGGACUAUUUCUC ((((((((....))))))))((((((..((((((.((((.(((((((.(((......)))..))))))).)).((((((.....))))))....)).))......))))..).))))).. ( -46.80) >consensus UGGCGUUGCAGGCAGUGAUAGAAGUGCUCCAGCUUCAGCGGAAGGAGUUGCAGCUUGGGGAACUUCUUUACCAGCAAGUUUGUGGCCUUGAGCACGGCCAUCAGGCUGGGAUUGUUCCGC ..(((..((((........((((((.((((((((.((((.......)))).))).))))).))))))...((....(((((((((((........))))).)))))).)).))))..))) (-34.54 = -36.14 + 1.60)

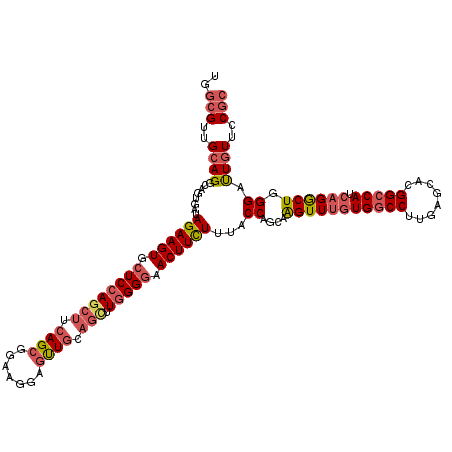
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:07 2006