
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,107,510 – 23,107,611 |
| Length | 101 |
| Max. P | 0.811364 |

| Location | 23,107,510 – 23,107,611 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.36 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -18.61 |
| Energy contribution | -16.23 |
| Covariance contribution | -2.38 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
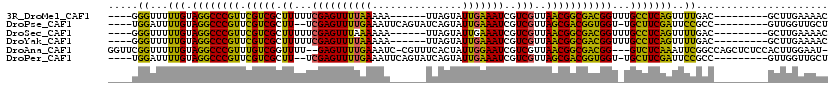
>3R_DroMel_CAF1 23107510 101 - 27905053 ----GGGUUUUUGUAGGCCCGUUCGUCGCUUUUUCGAGUUUUAAAAA------UUAGUAUUGAAAUCGUCGUUAACGGCGACGGUUUGCCUCAGUUUUGAC---------GCUUGAAAAC ----((((((....))))))..........(((((((((.((((((.------...(((....(((((((((.....)))))))))))).....)))))).---------))))))))). ( -30.10) >DroPse_CAF1 7394 104 - 1 ----UGGAUUUUGUAGGCCCGUUCGUCGCUU--UCGAGUUUUGAAAUUCAGUAUCAGUAUUGAAAUCGUCGUUAGCGACGGUGGU-UGCUUCGAUUCCGCC---------GUUGGUUGCU ----.(((..(((.((((....((((((((.--.(((....(((..(((((((....))))))).))))))..))))))))....-.))))))).)))((.---------.......)). ( -29.20) >DroSec_CAF1 10527 101 - 1 ----GGGUUUUUGUAGGCCCGUUCGUCGCUUUUUCGAGUUUAAAAAA------UUAGUAUUGAAAUCGUCGUUAACGGCGACGGUUUGCCUCAGUUUUGAC---------GCUUGAAAAC ----((((((....))))))..........(((((((((((((((..------...(((....(((((((((.....)))))))))))).....)))))).---------))))))))). ( -29.00) >DroYak_CAF1 10792 101 - 1 ----GGGUUUUUGUAGGCCCGUUCGUCGCUUUUUCGAGUUUUAAAAA------UUAGUAUUGAAAUCGUCGUUAACGGCGACGGUUUGCCUCAGUUUUGAC---------GCUUGAAAAC ----((((((....))))))..........(((((((((.((((((.------...(((....(((((((((.....)))))))))))).....)))))).---------))))))))). ( -30.10) >DroAna_CAF1 10781 113 - 1 GGUUCGGUUUUUGUAGGCCCGUUUGUCGGUUUU--GAGUUUUGAAAUC-CGUUUCACUAUUGGAAUCGUCGUUAACGGCGACGG---GUCUCAAAUUCGGCCAGCUCUCCACUUGGAAU- (((.(((..((((.(((((((((....(((((.--.(....(((((..-..)))))...)..)))))((((....)))))))))---)))))))).)))))).....(((....)))..- ( -38.10) >DroPer_CAF1 7368 104 - 1 ----UGGAUUUUGUAGGCCCGUUCGUCGCUU--UCGAGUUUUGAAAUUCAGUAUCAGUAUUGAAAUCGUCGUUAGCGACGGUGGU-UGCUUCGAUUCCGCC---------GUUGGUUGCU ----.(((..(((.((((....((((((((.--.(((....(((..(((((((....))))))).))))))..))))))))....-.))))))).)))((.---------.......)). ( -29.20) >consensus ____GGGUUUUUGUAGGCCCGUUCGUCGCUUUUUCGAGUUUUAAAAA______UCAGUAUUGAAAUCGUCGUUAACGGCGACGGU_UGCCUCAAUUUCGAC_________GCUUGAAAAC .....((...(((.((((((((.(((((.((...((((((((((...............)))))))..)))..)))))))))))...)))))))..))...................... (-18.61 = -16.23 + -2.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:04 2006