
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,106,104 – 23,106,289 |
| Length | 185 |
| Max. P | 0.999227 |

| Location | 23,106,104 – 23,106,217 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23106104 113 - 27905053 UUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU-UUUUUUUUUAAACCAUCUGCCAAGUGAAUUUCCAGUGGGCAGAAGUA----UACAUAUCAAACGG (((((...((((((((((....)))))))))).((((((.....))))))..-...............((((((.(.((......)).).))))))....----......)))))... ( -31.20) >DroSec_CAF1 9115 111 - 1 UUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU-UUUUUU--UUAACCAUCUGCCAAGUGAACUUCCAGUGGGCAGAAGUA----UACAUAUCAAACGG (((((...((((((((((....)))))))))).((((((.....))))))..-......--.......((((((.(.((......)).).))))))....----......)))))... ( -31.20) >DroSim_CAF1 10932 110 - 1 UUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU-UUU-UU--UUAACCAUCUGCCAAGUGAACUUCCAGUGGGCAGAAGUA----UACAUAUCAAACGG (((((...((((((((((....)))))))))).((((((.....))))))..-...-..--.......((((((.(.((......)).).))))))....----......)))))... ( -31.20) >DroEre_CAF1 9391 113 - 1 UUUGACACAAAAAGUCCA-AACUGGAUUUUUUCGCUGGCAGAUAGCCAGCAU-UUU-UU--UUAACCAUCUGCCAAGUGAAUUUCGGGUGGGUUGGAGUAUAUGUACAUAUCAAACGG ((..((..(((((((((.-....))))))))).((((((.....))))))..-...-..--....(((((((............)))))))))..))(((....)))........... ( -30.80) >DroYak_CAF1 9385 115 - 1 UUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCACAUAGCCAGCAUAUUU-UU--UUAACCAUCUGCCAACUGAAUUUCGGGUGGGUUGAAGUUUCUGUACAUAUCGAACGG ((..((..((((((((((....)))))))))).((((((.....))))))......-..--....(((((((............)))))))))..))....((((.(.....).)))) ( -29.00) >consensus UUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU_UUU_UU__UUAACCAUCUGCCAAGUGAAUUUCCAGUGGGCAGAAGUA____UACAUAUCAAACGG (((((...((((((((((....)))))))))).((((((.....))))))..................((((((.(.((......)).).))))))..............)))))... (-27.52 = -27.36 + -0.16)

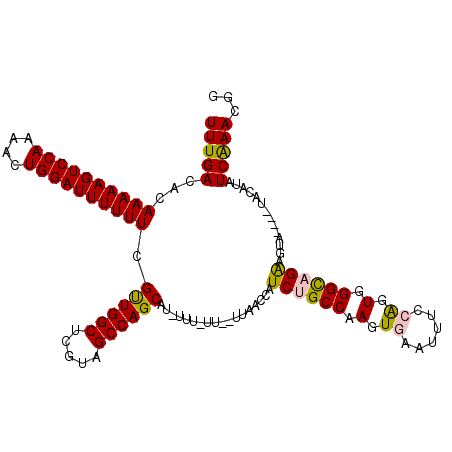
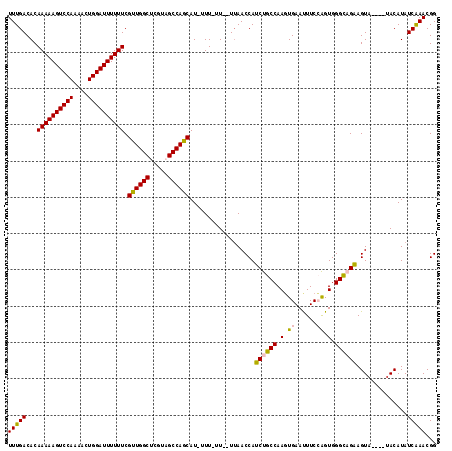
| Location | 23,106,140 – 23,106,252 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23106140 112 - 27905053 -----AACCGAAACCUAACCGAAUCCACGACUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU-UUUUUUUUUAAACCAUCUGCCAAGU -----...................................((((.((.((((((((((....)))))))))).((((((.....))))))..-.................)).)))). ( -19.30) >DroSec_CAF1 9151 110 - 1 -----AACCGAAACCCAACCGAAUCCAAGUCUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU-UUUUUU--UUAACCAUCUGCCAAGU -----....((((.......(((((((.((.......((((((......)))))).....)))))))))))))((((((.....))))))..-......--................. ( -20.11) >DroSim_CAF1 10968 109 - 1 -----AACCGAAACCCAACCGAAUCCACGACUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU-UUU-UU--UUAACCAUCUGCCAAGU -----...................................((((.((.((((((((((....)))))))))).((((((.....))))))..-...-..--.........)).)))). ( -19.30) >DroEre_CAF1 9431 113 - 1 CCGAAAACCGAAAGAUAACCGAAUCCACGACUGCUUGACUUUUGACACAAAAAGUCCA-AACUGGAUUUUUUCGCUGGCAGAUAGCCAGCAU-UUU-UU--UUAACCAUCUGCCAAGU ..((((..(....)......(((((((.(..((...(((((((......)))))))))-..))))))))))))((((((.....))))))..-...-..--................. ( -26.90) >DroYak_CAF1 9425 115 - 1 CCGAAAACCGAAAGAUAACCGAAUCCACGAUUGUCUGUCUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCACAUAGCCAGCAUAUUU-UU--UUAACCAUCUGCCAACU ........(((((((((..(((........)))..)))))))))....((((((((((....)))))))))).((((((.....))))))......-..--................. ( -27.50) >consensus _____AACCGAAACCUAACCGAAUCCACGACUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAU_UUU_UU__UUAACCAUCUGCCAAGU .........................................(((.((.((((((((((....)))))))))).((((((.....))))))....................)).))).. (-19.30 = -19.14 + -0.16)

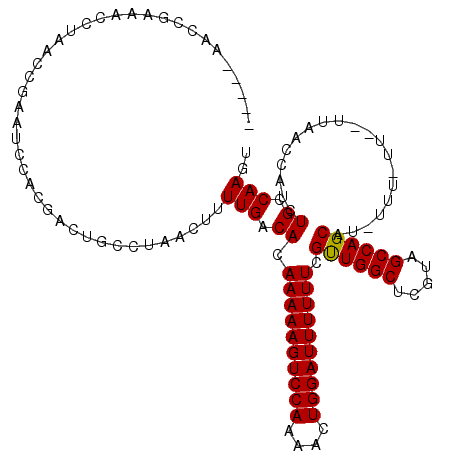

| Location | 23,106,179 – 23,106,289 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.38 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -21.82 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23106179 110 - 27905053 AGUAAAUAUCGAGUUGGUGGUGGAUUCUGAAGACGAA-------AACCGAAACCUAACCGAAUCCACGACUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGG ...........((..(((.((((((((..........-------...............)))))))).)))..)).....(..((..((((((((((....)))))))))).))..) ( -26.01) >DroSec_CAF1 9188 110 - 1 AGUAAAUAUCGAGUUGGUGGUGGAUUCUGAAGACGAA-------AACCGAAACCCAACCGAAUCCAAGUCUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGG ..........(((((((..((((((((..........-------...............)))))))...)..)).)))))(..((..((((((((((....)))))))))).))..) ( -24.91) >DroSim_CAF1 11004 110 - 1 AGUAAAUAUCGAGUUGGUGGUGGAUUCUGAAGACGAA-------AACCGAAACCCAACCGAAUCCACGACUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGG ...........((..(((.((((((((..........-------...............)))))))).)))..)).....(..((..((((((((((....)))))))))).))..) ( -26.01) >DroEre_CAF1 9467 116 - 1 AGUAAAUAUCGAGUUGGUGGUGGAUUCUGAAGACGAAAGCCGAAAACCGAAAGAUAACCGAAUCCACGACUGCUUGACUUUUGACACAAAAAGUCCA-AACUGGAUUUUUUCGCUGG ..((((..((((((.(((.((((((((....(.(....).)......(....)......)))))))).)))))))))..)))).((((((((((((.-....))))))))).).)). ( -34.30) >DroYak_CAF1 9462 117 - 1 AGUAAAUAUCGAGUUGGUGGUGGAUUCUGAAGACGAAUGCCGAAAACCGAAAGAUAACCGAAUCCACGAUUGUCUGUCUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGG ......((((...(((((..(((((((.......)))).)))...)))))..)))).......((((((.((((........))))..(((((((((....)))))))))))).))) ( -29.50) >consensus AGUAAAUAUCGAGUUGGUGGUGGAUUCUGAAGACGAA_______AACCGAAACCUAACCGAAUCCACGACUGCCUAACUUUUGACACAAAAAGUCCAAAACUGGAUUUUUUCGUUGG .((........((..(((.((((((((................................)))))))).)))..))........))..((((((((((....))))))))))...... (-21.82 = -21.90 + 0.08)

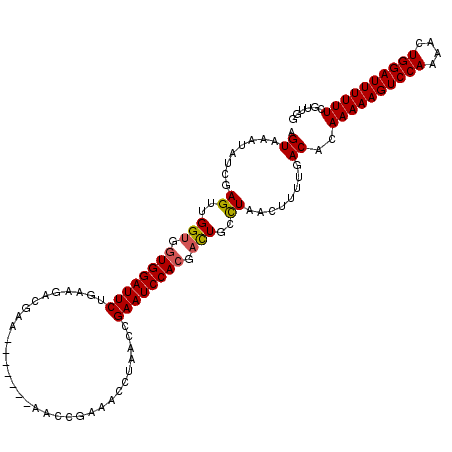
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:02 2006