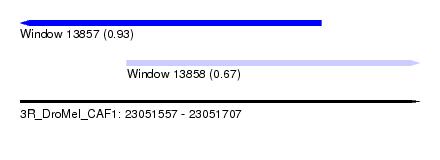
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,051,557 – 23,051,707 |
| Length | 150 |
| Max. P | 0.932745 |

| Location | 23,051,557 – 23,051,670 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -22.59 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23051557 113 - 27905053 AGUUUCGCCGGCAGCAGGCCAUUACGGCUGCAGCUAUCGCCGCUGCGUUUGCAGCAGAUGCUGCUUCAGUU------GCAGUUACAGUUGCAGUUGCUUCCUCAUCGUACUGAAACCCA .(((((...(((.....)))....((((((((((((((...(((((....))))).)))(((((.......------)))))...)))))))))))...............)))))... ( -39.90) >DroSec_CAF1 53352 94 - 1 AGUUUCGC-GGCAGCAGCCCAUUACUG---------------CUGCGUUCGCAGU---UGCUGCUGCAGUU------GCAGUUACAGUUGCAGUUGCUUCCUCAUCGCCAUGAAAUGCA .((((((.-(((((((((.....((((---------------(.......)))))---.))))))(((..(------((((......)))))..))).........))).))))))... ( -35.40) >DroSim_CAF1 36847 100 - 1 AGUUUCGC-GGCAGCAGCCCGUUACUG---------------CUGCGUUCGCAGU---UGCUGCUGCAGUUGCAGUUGCAGUUACAGUUGCAGUUGCUUCCUCAUCGCACUGAAAUCCA .(((((((-(((((((((.....((((---------------(.......)))))---.))))))))....(((..(((((......)))))..)))........)))...)))))... ( -36.50) >consensus AGUUUCGC_GGCAGCAGCCCAUUACUG_______________CUGCGUUCGCAGU___UGCUGCUGCAGUU______GCAGUUACAGUUGCAGUUGCUUCCUCAUCGCACUGAAAUCCA .(((((((.((.((((((........................(((((...(((((....))))))))))........((((......)))).)))))).)).....))...)))))... (-22.59 = -22.60 + 0.01)


| Location | 23,051,597 – 23,051,707 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -9.37 |
| Energy contribution | -9.73 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23051597 110 + 27905053 GC------AACUGAAGCAGCAUCUGCUGCAAACGCAGCGGCGAUAGCUGCAGCCGUAAUGGCCUGCUGCCGGCGAAACUGUCGCCAGACAAAUCAAAAGAAAUCGCUCUGGAAUAC ..------.......(((((....))))).....(((.((((((.((.((((((.....)).)))).)).(((((.....)))))................)))))))))...... ( -38.00) >DroSec_CAF1 53392 91 + 1 GC------AACUGCAGCAGCA---ACUGCGAACGCAG---------------CAGUAAUGGGCUGCUGCC-GCGAAACUGUCGCCAGACAAAUCAAAAGAAAUCGCUCUGGAAUAC ..------....((((((((.---.((((....))))---------------((....)).)))))))).-((((...((((....))))..((....))..)))).......... ( -30.30) >DroSim_CAF1 36887 97 + 1 GCAACUGCAACUGCAGCAGCA---ACUGCGAACGCAG---------------CAGUAACGGGCUGCUGCC-GCGAAACUGUCGCCAGACAAAUCAAAAGAAAUCGCUCUGGAAUAC ((....))....((((((((.---(((((.......)---------------)))).....)))))))).-((((...((((....))))..((....))..)))).......... ( -31.50) >DroEre_CAF1 59078 83 + 1 --------AGCAGCAGCGAUA---UCU------GCAG---------------CAGUAAUAGGUUGCAGCC-GCGAAUCUGUCGCCAGACAAAUCAAAAGAAAUCGCUCUGGAAUAC --------..(((.((((((.---(((------((.(---------------(.((((....)))).)).-))((...((((....))))..))...))).)))))))))...... ( -24.80) >DroYak_CAF1 58449 83 + 1 --------AACUGCAGCAGCA---UCU------GCAG---------------CAGUAAUAGGCUGCAGCC-GCGAAACUGUCGCCAGACAAAUCAAAAGAAAUCGCUCUGGAAUAC --------.(((((.((((..---.))------)).)---------------))))....(((....)))-((((...((((....))))..((....))..)))).......... ( -24.10) >DroAna_CAF1 58062 84 + 1 GA------AACCUCAGAA-AA---UCA------GCAG---------------CCAUAAGUCUCAGCCGCC-ACGAAACUGUCGCCAGACAAAUCAAACCAAUUCGCUGUGGGACAC ..------..((.(((..-((---(..------((.(---------------(...........)).)).-..((...((((....))))..))......)))..))).))..... ( -12.40) >consensus GC______AACUGCAGCAGCA___UCU______GCAG_______________CAGUAAUGGGCUGCUGCC_GCGAAACUGUCGCCAGACAAAUCAAAAGAAAUCGCUCUGGAAUAC ...............(((((.........................................)))))..((.((((...((((....))))..((....))..))))...))..... ( -9.37 = -9.73 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:45 2006