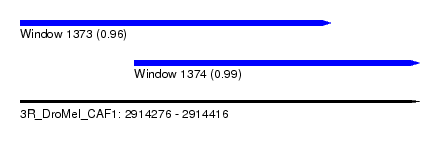
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,914,276 – 2,914,416 |
| Length | 140 |
| Max. P | 0.986951 |

| Location | 2,914,276 – 2,914,385 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2914276 109 + 27905053 GCGAGGGUGAGGGCGCUGAGGAGUACUAAGCGUCACGCCA--C-UUCAA--CGCUCGAUGGGAGCGUCAUUGGUGGGCGGGGU-AACCGUCGAAAUCAGUGUUUACGC-----UUCCAAU (((((((((..((((((.((.....)).)))))).)))).--)-)....--)))....((((((((((((((((.(((((...-..)))))...)))))))...))))-----))))).. ( -48.20) >DroVir_CAF1 5115 118 + 1 GUGAGGGCGAGGGUGCCGAGGAGUACUAAACACG-CUCCACACUUCCAGCCCGCACAGAGCAAGCGUCAUUGGUGGGCGUUGA-GCACGUCGAAACCAAUGCCCACGCAUUGCCUUCGUU (((.(((((..((((....(((((.........)-)))).))))..).)))).)))((.(((((((((((((((.(((((...-..)))))...)))))))...)))).))))))..... ( -46.00) >DroSec_CAF1 5098 109 + 1 GCGAGGGAGAGGGUGCUGAGGAGUACUAAGCGUCGCGCCA--C-UCCAA--CGCUCGAUGGGAGCGUCAUUGGUGGGCGGGGA-AACCGUCGAAAUCAUUGUUUACGC-----UUCCAAU (((..((((..(((((.((.(.........).))))))).--)-)))..--)))....(((((((((((.((((.(((((...-..)))))...)))).))...))))-----))))).. ( -46.50) >DroEre_CAF1 5171 109 + 1 GCGAGGGCGAGGGUGCUGAGGAGUACUAAGCGUGCCGCCA--C-UCCAA--CGCUCGACUGGAGCGUCAUUGGUGGGCGGGAA-AACCGUCGAAAUCAGUGUUUACGC-----UUCCAAU ..(..((((..((..((((...((((.....))))(((((--.-....(--(((((.....))))))...)))))(((((...-..)))))....))))..))..)))-----)..)... ( -42.20) >DroWil_CAF1 6097 108 + 1 GCGAGGGUGAGGGAGCUGAGGAGUACUAGAGAUCC--CCA--C-AACAU--AAAUGGGAUGAAGCGUCACUAGUGGGCGUCUAAAGACGUCGAAACUAAUGAUUACGC-----UUCAUUU ......(((.((((.((.((.....))..)).)))--)))--)-.....--.....(((((((((((((.((((.((((((....))))))...)))).)))...)))-----))))))) ( -37.70) >DroYak_CAF1 4974 109 + 1 GUGAGGGCGAGGGUGCUGAGGAGUACUAAGCGUGCCGCCA--C-UCCAA--CGCUCGAUGGGAGCGUCAUUGGUGGGCGGGAA-AACCGUCGAAAUCAGUGUUUACGC-----UUCCAAU (.(((((((.(.(((((.((.....)).))))).))))).--)-)))..--.......((((((((((((((((.(((((...-..)))))...)))))))...))))-----))))).. ( -46.10) >consensus GCGAGGGCGAGGGUGCUGAGGAGUACUAAGCGUCCCGCCA__C_UCCAA__CGCUCGAUGGGAGCGUCAUUGGUGGGCGGGGA_AACCGUCGAAAUCAGUGUUUACGC_____UUCCAAU (((.(((((..((((((....))))))...)))....))............(((((.....))))).(((((((.(((((......)))))...)))))))....)))............ (-23.44 = -24.20 + 0.76)

| Location | 2,914,316 – 2,914,416 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.45 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2914316 100 + 27905053 CUUCAACGCUCGAUGGGAGCGUCAUUGGUGGGCGGGGU-AACCGUCGAAAUCAGUGUUUACGCUUCCAAUCGCAACAAAAAA------UUCACUG---CAACACUGAAAA .((((......((((((((((((((((((.(((((...-..)))))...)))))))...)))))))).)))(((........------.....))---).....)))).. ( -32.42) >DroSec_CAF1 5138 100 + 1 CUCCAACGCUCGAUGGGAGCGUCAUUGGUGGGCGGGGA-AACCGUCGAAAUCAUUGUUUACGCUUCCAAUCGCAACAAAAAA------UUCACUA---CAACACUGAAAA ..........((((((((((((((.((((.(((((...-..)))))...)))).))...)))))))).))))..........------.......---............ ( -27.30) >DroSim_CAF1 5174 100 + 1 CUCCAACGCUCGAUGGGAGCGUCAUUGGUGGGCGGGGA-AACCGUCGAAAUCAGUGUUUACGCUUCCAAUCGCAACAAAAAA------UUCACUA---CAACACUGAAAA ..........(((((((((((((((((((.(((((...-..)))))...)))))))...)))))))).))))..........------.......---............ ( -30.50) >DroEre_CAF1 5211 100 + 1 CUCCAACGCUCGACUGGAGCGUCAUUGGUGGGCGGGAA-AACCGUCGAAAUCAGUGUUUACGCUUCCAAUCGCAACAAAAAA------UUCACUG---CAACACUGUAAA ..........(((.(((((((((((((((.(((((...-..)))))...)))))))...)))).)))).)))..........------.....((---((....)))).. ( -28.20) >DroWil_CAF1 6135 108 + 1 CAACAUAAAUGGGAUGAAGCGUCACUAGUGGGCGUCUAAAGACGUCGAAACUAAUGAUUACGCUUCAUUUAAAAAAAAAAAACAAAUGUUCACUGUUUCUACACUGAA-- ...........(((((((((((((.((((.((((((....))))))...)))).)))...))))))))))..................((((.(((....))).))))-- ( -29.20) >DroYak_CAF1 5014 100 + 1 CUCCAACGCUCGAUGGGAGCGUCAUUGGUGGGCGGGAA-AACCGUCGAAAUCAGUGUUUACGCUUCCAAUCGCAACAAAAAA------UUCACUG---CAACACUGUAAA ..........(((((((((((((((((((.(((((...-..)))))...)))))))...)))))))).))))..........------.....((---((....)))).. ( -31.40) >consensus CUCCAACGCUCGAUGGGAGCGUCAUUGGUGGGCGGGGA_AACCGUCGAAAUCAGUGUUUACGCUUCCAAUCGCAACAAAAAA______UUCACUG___CAACACUGAAAA ..........(((((((((((((((((((.(((((......)))))...)))))))...)))))))).))))...................................... (-27.86 = -27.45 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:04 2006