
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,026,966 – 23,027,062 |
| Length | 96 |
| Max. P | 0.882584 |

| Location | 23,026,966 – 23,027,062 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23026966 96 + 27905053 -------CCUUGUGUUUCCUUUUCGCUUUCGUGCCCAGUUC-----GCGG-----A----CUUGGC---GCAACAUUUCAACCUGCCACUGCGGUGGCAGCAUUGAUGGGCGUAUACAUG -------...(((((........((((...(((((.((((.-----...)-----)----)).)))---))..((((.....(((((((....)))))))....)))))))).))))).. ( -33.60) >DroVir_CAF1 34986 90 + 1 -----------------GCUUUUCGCUUUCGUGCCCAGUCC-----GC-A----AACUCCCUUCGG---GCAACAUUUCAACCUGCCACUGCGGUGGCAGCAUUGAUGGGCGUAUACAUG -----------------((.....))......(((((((((-----(.-(----(......)))))---))......((((.(((((((....)))))))..)))))))))......... ( -30.50) >DroGri_CAF1 38592 105 + 1 ---------------CUGAUUUUCGCUUUCGUGCCCAGUGCCGUCCGCGGAGUGGACUCCCUUCGGGGAGCAACAUUUCAACCUGCCACUGCGGUGGCAGCAUUGAUGGGCGUAUACAUG ---------------.................(((((.(((.((((((...))))))((((....))))))).....((((.(((((((....)))))))..)))))))))......... ( -42.50) >DroWil_CAF1 35717 103 + 1 GACUUUGGCUUGGG-UUCCUUUUCGCUUUCGUGCCCAGUCU-----GCAA----AA----GUUGGC---GCAACAUUUCAACCUGCCACUGUGGUGGCAGCAUUGAUGGGCGUAUACAUG ((((((.(((((((-(................))))))...-----)).)----))----))).((---((..((((.....(((((((....)))))))....)))).))))....... ( -31.99) >DroMoj_CAF1 38342 92 + 1 -----------------ACAUUUCGCUUUCGUGCCCAGACC----CGCGA----AACUCCCUUCGG---GCAACAUUUCAACCUGCCACUGCGGUGGCAGCAUUGAUGGGCGUAUACAUG -----------------...............(((((...(----(.(((----(......)))))---).......((((.(((((((....)))))))..)))))))))......... ( -28.20) >DroPer_CAF1 31686 96 + 1 -------CCUUGUGUUUCCAUUUCGCUUUCGUGCCCAGUCC-----GCGG-----A----CUUGGC---GCAACAUUUCAACCUGCCACUGCGGUGGCAGCAUUGAUGGGCGUAUACAUG -------...(((((.((((((..((....(((((.((((.-----...)-----)----)).)))---))............((((((....))))))))...))))))...))))).. ( -36.00) >consensus _______________UUCCUUUUCGCUUUCGUGCCCAGUCC_____GCGA____AA____CUUCGC___GCAACAUUUCAACCUGCCACUGCGGUGGCAGCAUUGAUGGGCGUAUACAUG ................................(((((.........((.....................))......((((.(((((((....)))))))..)))))))))......... (-22.77 = -22.77 + 0.00)

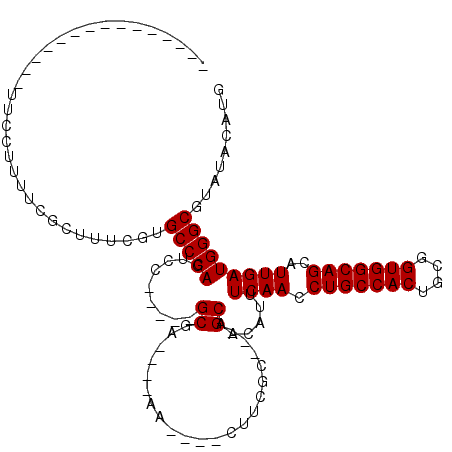

| Location | 23,026,966 – 23,027,062 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23026966 96 - 27905053 CAUGUAUACGCCCAUCAAUGCUGCCACCGCAGUGGCAGGUUGAAAUGUUGC---GCCAAG----U-----CCGC-----GAACUGGGCACGAAAGCGAAAAGGAAACACAAGG------- ..(((....((((((((((.(((((((....))))))))))))....((((---(.....----.-----.)))-----))..))))).(....)......(....))))...------- ( -31.80) >DroVir_CAF1 34986 90 - 1 CAUGUAUACGCCCAUCAAUGCUGCCACCGCAGUGGCAGGUUGAAAUGUUGC---CCGAAGGGAGUU----U-GC-----GGACUGGGCACGAAAGCGAAAAGC----------------- .........((((((((((.(((((((....))))))))))))..(((.((---((....)).)).----.-))-----)...))))).(....)........----------------- ( -31.50) >DroGri_CAF1 38592 105 - 1 CAUGUAUACGCCCAUCAAUGCUGCCACCGCAGUGGCAGGUUGAAAUGUUGCUCCCCGAAGGGAGUCCACUCCGCGGACGGCACUGGGCACGAAAGCGAAAAUCAG--------------- .........((((((((((.(((((((....)))))))))))).....((((..(((...((((....)))).)))..)))).))))).(....)..........--------------- ( -40.10) >DroWil_CAF1 35717 103 - 1 CAUGUAUACGCCCAUCAAUGCUGCCACCACAGUGGCAGGUUGAAAUGUUGC---GCCAAC----UU----UUGC-----AGACUGGGCACGAAAGCGAAAAGGAA-CCCAAGCCAAAGUC .........((((((((((.(((((((....))))))))))))....((((---(.....----..----.)))-----))..))))).(....)......((..-......))...... ( -29.40) >DroMoj_CAF1 38342 92 - 1 CAUGUAUACGCCCAUCAAUGCUGCCACCGCAGUGGCAGGUUGAAAUGUUGC---CCGAAGGGAGUU----UCGCG----GGUCUGGGCACGAAAGCGAAAUGU----------------- .........((((((((((.(((((((....))))))))))))......((---(((.........----...))----))).))))).(....)........----------------- ( -31.80) >DroPer_CAF1 31686 96 - 1 CAUGUAUACGCCCAUCAAUGCUGCCACCGCAGUGGCAGGUUGAAAUGUUGC---GCCAAG----U-----CCGC-----GGACUGGGCACGAAAGCGAAAUGGAAACACAAGG------- ..(((...(((...(((((.(((((((....))))))))))))....(((.---(((.((----(-----(...-----.)))).))).)))..)))....(....))))...------- ( -35.00) >consensus CAUGUAUACGCCCAUCAAUGCUGCCACCGCAGUGGCAGGUUGAAAUGUUGC___CCCAAG____UU____CCGC_____GGACUGGGCACGAAAGCGAAAAGGAA_______________ .........((((((((((.(((((((....))))))))))))...(........)...........................))))).(....)......................... (-23.15 = -23.15 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:39 2006