
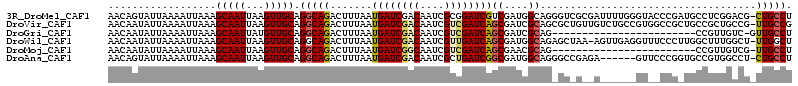
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,017,237 – 23,017,350 |
| Length | 113 |
| Max. P | 0.993680 |

| Location | 23,017,237 – 23,017,350 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.29 |
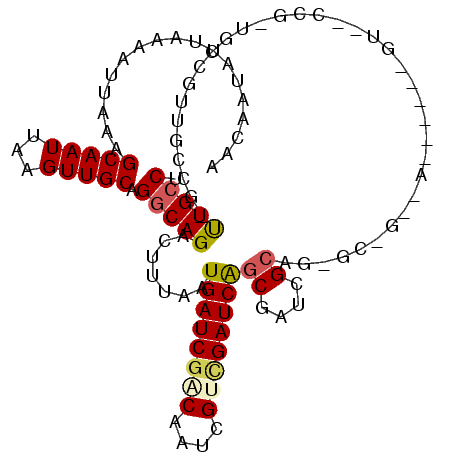
| Mean single sequence MFE | -34.13 |
| Consensus MFE | -18.81 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23017237 113 + 27905053 AACAGUAUUAAAAUUAAAGCAAUUAAGUUGCAGGCAGACUUUAAUGAUCGACAAUCGCGGAUCGUCGAUGGCAGGGUCGCGAUUUUGGGUACCCGAUGCCUCGGACG-CUGCCU .....(((((((......(((((...))))).(.....))))))))((((((.(((...))).))))))(((((.(((.(((..((((....))))....)))))).-))))). ( -34.20) >DroVir_CAF1 20946 113 + 1 AACAAUAUUAAAAUUAAAGCAAUUAAGUUGCAGGCAGACUUUAAUGAUCGACAAUCGUCGAUCAGCGAUCGCAGCGCUGUUGUCUGCCGUGGCCGCUGCCGCUGCCG-UUGCCG ..................(((((......((((((((((......(((((((....)))))))((((.......))))...)))))))(((((....)))))))).)-)))).. ( -40.30) >DroGri_CAF1 26783 89 + 1 AACAAUAUUAAAAUUAAAGCAAUUAUGUUGCAGGCAGACUUUAAUGAUCGACAAUCGUCGAUCAGCGAUCGCAG------------------------CCGUUGUC-GUUGCCU ..................(((((...)))))(((((........((((((((....))))))))(((((.((..------------------------..)).)))-))))))) ( -26.90) >DroWil_CAF1 25014 112 + 1 AACAAUAUUAAAAUUAAAGCAAUUAAGUUGCAGGCAGACUUUAAUGAUCGACAAUCGUUGAUCAGCGAUGGCAGAGCUAA-AGUUGAGGUUUCCCUUGGCUUUGGCU-UUGGCU .................(((......(((((((.....))....((((((((....)))))))))))))..(((((((((-(((..(((....)))..)))))))))-)))))) ( -40.10) >DroMoj_CAF1 25887 89 + 1 AACAAUAUUAAAAUUAAAGCAAUUAAGUUGCAGGCAGACUUUAAUGAUCGGCAAUCGUCGAUCAGCGAACGCAG------------------------CCGUUGUCG-UUGCCU ..................(((((...)))))((((((((.....((((((((....))))))))((....))..------------------------.....))).-.))))) ( -24.70) >DroAna_CAF1 19681 107 + 1 AACAGUAUUAAAAUUAAAGCAAUUAAGUUGCAGGCAGACUUUAAUGAUCGACAAUCGCUGAUCGGCGAUGGCAGGGCCGAGA------GUUCCCGGUGCCGUGGCCU-CUGCCU ..................(((((...)))))(((((((.......(((((........)))))(((.(((((((((.(....------)..)))..)))))).))))-)))))) ( -38.60) >consensus AACAAUAUUAAAAUUAAAGCAAUUAAGUUGCAGGCAGACUUUAAUGAUCGACAAUCGUCGAUCAGCGAUCGCAG_GC_G__A______GU__CCG_UGCCGUUGCCG_UUGCCU ..................(((((...))))).(((((.......((((((((....))))))))((....))....................................))))). (-18.81 = -18.95 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:33 2006