
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,997,390 – 22,997,506 |
| Length | 116 |
| Max. P | 0.995142 |

| Location | 22,997,390 – 22,997,481 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.36 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -11.73 |
| Energy contribution | -11.92 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
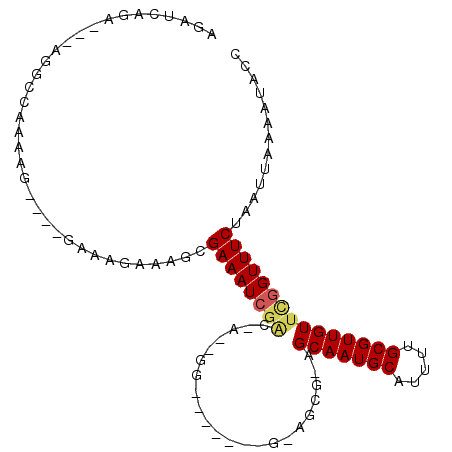
>3R_DroMel_CAF1 22997390 91 - 27905053 AUCGAAAUCGAA-AUCGACCAGCGAGCAGCCUUGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAGCC-----GG-----CCGAAAC .(((....))).-.(((.((.(((((((((...((((......)))))))))))(((.............))).......)).-----))-----.)))... ( -23.62) >DroPse_CAF1 1339 94 - 1 AGCGAAAUCGGGAAUUGG-------GAAUCG-AGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACCAAAAAGAGCCGAGCCGAGCCAGCCGACAC .......((((.((((((-------((((((-((((((((.....)))))))))))))))))))).............(.((((...)).)))..))))... ( -30.70) >DroGri_CAF1 3703 70 - 1 AGCAAAAC--------GG-------CCAGAG-AGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACCAAAAAAA---------------CCGAAA- .......(--------((-------.....(-((((((((.....)))))))))(((.............))).......---------------)))...- ( -15.12) >DroEre_CAF1 3634 77 - 1 AUCGAAAUCGACCA--------------GCG-AGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAGCC-----GG-----CCGAAAC .......(((.((.--------------(((-((((((((.....)))))))))(((.............))).......)).-----))-----.)))... ( -23.02) >DroAna_CAF1 1552 80 - 1 AGCGAAAUCGACCG--------------GCG-AGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAGCC-----AGAGCCGCCG--AC .......(((..((--------------(((-((((((((.....)))))))))(((((...................)))))-----...)))).))--). ( -24.91) >DroPer_CAF1 1375 89 - 1 AGCGAAAUCGGGAAUUGG-------GAAUCG-AGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACCAAAAAGAGCC-----GAGCCAGCCGACAC .((....((((.((((((-------((((((-((((((((.....)))))))))))))))))))).......(.....)..))-----))....))...... ( -30.30) >consensus AGCGAAAUCGAC_A__GG_______G_AGCG_AGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAGCC_____GG_____CCGAAAC ...((((((((......................(((((((.....))))))))))))))).......................................... (-11.73 = -11.92 + 0.19)



| Location | 22,997,409 – 22,997,506 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.52 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22997409 97 - 27905053 AGAUCAGA---AGGCCAGAAG----GAAAGAAAUCGAAAUCGAA-AUCGACCAGCGAGCAGCCUUGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACC .(((.(((---((.((....(----(...((..(((....))).-.))..))...(((((((...((((......)))))))))))))))))).)))........ ( -23.10) >DroPse_CAF1 1368 93 - 1 AGAUCAGAAAAAGGCCCAAAG----CAGAGAAAGCGAAAUCGGGAAUUGG-------GAAUCG-AGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACC ..............(((...(----(.......))(....))))((((((-------((((((-((((((((.....))))))))))))))))))))........ ( -29.10) >DroEre_CAF1 3653 83 - 1 AGAUCAGA---AGGCCAGGAG----GAAAGAAAUCGAAAUCGACCA--------------GCG-AGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACC .(((.(((---((.((.....----........(((....)))...--------------..(-((((((((.....)))))))))))))))).)))........ ( -21.10) >DroMoj_CAF1 3345 86 - 1 GGAUCGAA---CGGCCGAAAGGCUCGAAAGAAAGCGAAAU--------GG-------CCAAAG-AGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACC ((((((((---(((((....)))((....))..(((((((--------((-------(.....-.))....))))))))..)))))))))).............. ( -27.40) >DroAna_CAF1 1574 83 - 1 AGAUCAGA---AGGCCAAAAG----GAAAGAAAGCGAAAUCGACCG--------------GCG-AGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACC .(((.(((---((.((....(----(...((........))..)).--------------..(-((((((((.....)))))))))))))))).)))........ ( -20.60) >DroPer_CAF1 1399 93 - 1 AGAUCAGAAAAAGGCCCAAAG----CAGAGAAAGCGAAAUCGGGAAUUGG-------GAAUCG-AGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACC ..............(((...(----(.......))(....))))((((((-------((((((-((((((((.....))))))))))))))))))))........ ( -29.10) >consensus AGAUCAGA___AGGCCAAAAG____GAAAGAAAGCGAAAUCGAC_A__GG_______G_AGCG_AGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACC ...................................((((((((......................(((((((.....)))))))))))))))............. (-12.36 = -12.52 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:21 2006