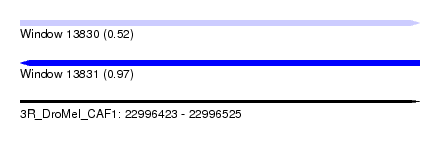
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,996,423 – 22,996,525 |
| Length | 102 |
| Max. P | 0.965508 |

| Location | 22,996,423 – 22,996,525 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516251 |
| Prediction | RNA |
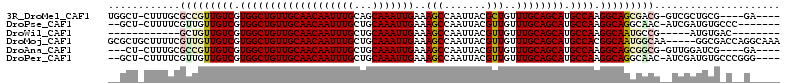
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22996423 102 + 27905053 ----UC----CGCAGCGAC-CGUCGCUGCCUUGGCAUGCUGCAAACAGCGUAAUUGGCUUUCAAUUUGCUGCAAAUUGUUGCAACAGCCACGACAACGGCGCAAAG-AGCCA ----..----.((((((..-...))))))......((((((....))))))...((((((....((((((((((....)))))...(((........)))))))))-))))) ( -37.00) >DroPse_CAF1 571 101 + 1 -------GGGCACAUCGAU-GUUGCCUGCCUUGGCAUGCUGCAAACAACGUAAUUGGCUUUCAAUUUGCAGCAAAUUGUUGCAACAGCCACGACAACAACGAAAAG-AGC-- -------.......(((.(-(((((.(((....))).))..........((...(((((......(((((((.....))))))).)))))..)))))).)))....-...-- ( -25.90) >DroWil_CAF1 3802 87 + 1 --------GUCACAU-----CGGCAUUGCCUUGGCAUGCUGCAAACAACGUAAUUGGCUUUCAAUUUGCAGCAAAUUGUUGCAACAGCCACGACAACAGC------------ --------(((....-----((((((.((....)))))))).............(((((......(((((((.....))))))).))))).)))......------------ ( -26.10) >DroMoj_CAF1 2502 107 + 1 UUUGCCUGGUCGCC-----UUGCCAUUGCCGUGGCAUGCUGCAAACAACGUAAUUGGCUUUCAAUUUGCAGCAAAUUGUUGCAACAGCCACGACAACAACGAAAAGCAGCGC ..((((((((.((.-----..))....)))).)))).(((((............(((((......(((((((.....))))))).)))))((.......))....))))).. ( -30.30) >DroAna_CAF1 467 99 + 1 ----UC----CGAUCCAAC-CGCCGCUGCCUUGGCAUGCUGCAAACAACGUAAUUGGCUUUCAAUUUGCAGCAAAUUGUUGCAACAGCCACGACAACGGCGCAAAG-AG--- ----..----.........-(((((.((.(.((((.(((((((((....((.....))......)))))))))...(((....))))))).).)).))))).....-..--- ( -28.20) >DroPer_CAF1 583 104 + 1 ----CCCGGGCACAUCGAU-GUUGCCUGCCUUGGCAUGCUGCAAACAACGUAAUUGGCUUUCAAUUUGCAGCAAAUUGUUGCAACAGCCACGACAACAACGAAAAG-AGC-- ----..........(((.(-(((((.(((....))).))..........((...(((((......(((((((.....))))))).)))))..)))))).)))....-...-- ( -25.90) >consensus _____C__G_CACAUCGA__CGUCACUGCCUUGGCAUGCUGCAAACAACGUAAUUGGCUUUCAAUUUGCAGCAAAUUGUUGCAACAGCCACGACAACAACGAAAAG_AGC__ ..........................((.(.((((.(((((((((....((.....))......)))))))))...(((....))))))).).))................. (-16.67 = -16.83 + 0.17)

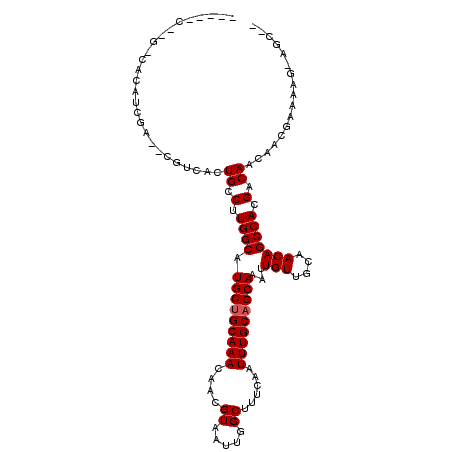
| Location | 22,996,423 – 22,996,525 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -29.17 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22996423 102 - 27905053 UGGCU-CUUUGCGCCGUUGUCGUGGCUGUUGCAACAAUUUGCAGCAAAUUGAAAGCCAAUUACGCUGUUUGCAGCAUGCCAAGGCAGCGACG-GUCGCUGCG----GA---- ..((.-....((((((((((..((((((((((((......(((((.(((((.....)))))..))))))))))))).)))).....))))))-).))).)).----..---- ( -40.40) >DroPse_CAF1 571 101 - 1 --GCU-CUUUUCGUUGUUGUCGUGGCUGUUGCAACAAUUUGCUGCAAAUUGAAAGCCAAUUACGUUGUUUGCAGCAUGCCAAGGCAGGCAAC-AUCGAUGUGCCC------- --((.-(...(((.(((((((.(((((((((((((((((((...))))))).....((((...)))).)))))))).)))).....))))))-).))).).))..------- ( -34.50) >DroWil_CAF1 3802 87 - 1 ------------GCUGUUGUCGUGGCUGUUGCAACAAUUUGCUGCAAAUUGAAAGCCAAUUACGUUGUUUGCAGCAUGCCAAGGCAAUGCCG-----AUGUGAC-------- ------------((.((((((.(((((((((((((((((((...))))))).....((((...)))).)))))))).)))).))))))))..-----.......-------- ( -28.50) >DroMoj_CAF1 2502 107 - 1 GCGCUGCUUUUCGUUGUUGUCGUGGCUGUUGCAACAAUUUGCUGCAAAUUGAAAGCCAAUUACGUUGUUUGCAGCAUGCCACGGCAAUGGCAA-----GGCGACCAGGCAAA ..((((((((..((..(((((((((((((((((((((((((...))))))).....((((...)))).)))))))).))))))))))..))))-----))).....)))... ( -42.70) >DroAna_CAF1 467 99 - 1 ---CU-CUUUGCGCCGUUGUCGUGGCUGUUGCAACAAUUUGCUGCAAAUUGAAAGCCAAUUACGUUGUUUGCAGCAUGCCAAGGCAGCGGCG-GUUGGAUCG----GA---- ---.(-((...((((((((((.(((((((((((((((((((...))))))).....((((...)))).)))))))).)))).))))))))))-...)))...----..---- ( -41.70) >DroPer_CAF1 583 104 - 1 --GCU-CUUUUCGUUGUUGUCGUGGCUGUUGCAACAAUUUGCUGCAAAUUGAAAGCCAAUUACGUUGUUUGCAGCAUGCCAAGGCAGGCAAC-AUCGAUGUGCCCGGG---- --((.-(...(((.(((((((.(((((((((((((((((((...))))))).....((((...)))).)))))))).)))).....))))))-).))).).)).....---- ( -34.50) >consensus __GCU_CUUUUCGCUGUUGUCGUGGCUGUUGCAACAAUUUGCUGCAAAUUGAAAGCCAAUUACGUUGUUUGCAGCAUGCCAAGGCAGCGACG__UCGAUGCG_C__G_____ ............(((((((((.(((((((((((((((((((...)))))))..(((.......)))..)))))))).)))).)))))))))..................... (-29.17 = -29.73 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:19 2006