
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,995,570 – 22,995,881 |
| Length | 311 |
| Max. P | 0.958515 |

| Location | 22,995,570 – 22,995,671 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -29.02 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22995570 101 + 27905053 UACCAGUCGGUUGUGGUUUGUAACUCCUCCAGGAGCUCUGUGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG ...(((.((((....((......((((....))))......))....))))))).(((((.(((((((((((...........))))))))))).))))). ( -35.10) >DroSec_CAF1 3200 92 + 1 UACCAGUCGGUUGUGGUUUGUAACUCCUCCAGGAGCUCUGUGCAGUUGC---------UGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG .((((........))))(((((.((((....)))).....))))).(((---------((.(((((((((((...........))))))))))).))))). ( -30.70) >DroSim_CAF1 3210 101 + 1 UACCAGUCGGUUGUGGUUUGCAACUCCUCCAGGAGCUCUGUGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG ...(((.((((......(((((.((((....)))).....)))))..))))))).(((((.(((((((((((...........))))))))))).))))). ( -36.80) >consensus UACCAGUCGGUUGUGGUUUGUAACUCCUCCAGGAGCUCUGUGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG .((((........))))..((((((((....)))((.....)).)))))......(((((.(((((((((((...........))))))))))).))))). (-29.02 = -29.80 + 0.78)


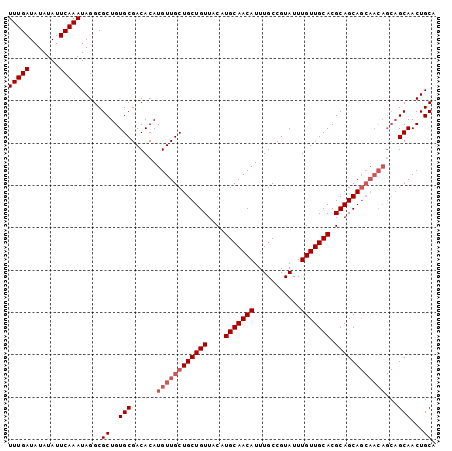
| Location | 22,995,570 – 22,995,671 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -27.70 |
| Energy contribution | -28.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22995570 101 - 27905053 CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCACAGAGCUCCUGGAGGAGUUACAAACCACAACCGACUGGUA .(((((((.(.(((((((...........))))))).).)))))))...((((...))))....((((((....))))))....((((........)))). ( -32.40) >DroSec_CAF1 3200 92 - 1 CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCA---------GCAACUGCACAGAGCUCCUGGAGGAGUUACAAACCACAACCGACUGGUA .(((((((.(.(((((((...........))))))).).))))---------))).........((((((....))))))....((((........)))). ( -28.60) >DroSim_CAF1 3210 101 - 1 CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCACAGAGCUCCUGGAGGAGUUGCAAACCACAACCGACUGGUA .(((((((.(.(((((((...........))))))).).)))))))......((((((.(.(((.....)))..).))))))..((((........)))). ( -33.90) >consensus CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCACAGAGCUCCUGGAGGAGUUACAAACCACAACCGACUGGUA (((((((....(((((((...........))))))).)))))))........((....))....((((((....))))))....((((........)))). (-27.70 = -28.03 + 0.33)



| Location | 22,995,610 – 22,995,711 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -29.43 |
| Energy contribution | -31.43 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.16 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22995610 101 + 27905053 UGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAA .((.(((((((((((((((((.((((....(......)....)))).))).)))))))))))))).((((...)))))).....(((((.......))))) ( -36.40) >DroSec_CAF1 3240 92 + 1 UGCAGUUGC---------UGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAA .((.(((((---------((((((((((((((...........)))))))))...)))))))))).((((...)))))).....(((((.......))))) ( -32.60) >DroSim_CAF1 3250 101 + 1 UGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAA .((.(((((((((((((((((.((((....(......)....)))).))).)))))))))))))).((((...)))))).....(((((.......))))) ( -36.40) >consensus UGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAA .((.(((((((((((......(((((((((((...........)))))))))))))))))))))).((((...)))))).....(((((.......))))) (-29.43 = -31.43 + 2.00)

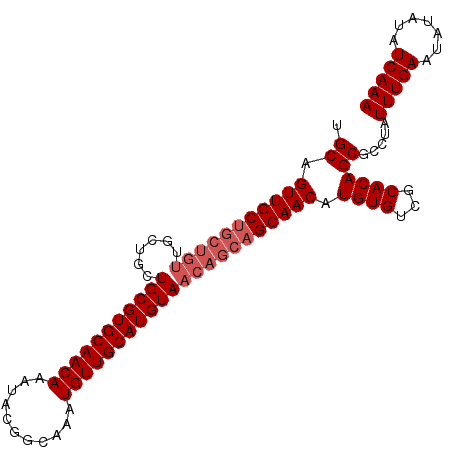
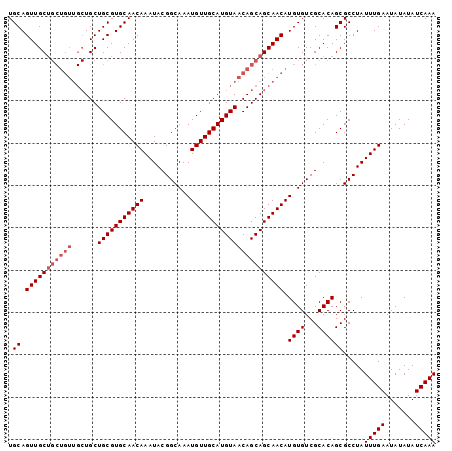
| Location | 22,995,610 – 22,995,711 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -29.43 |
| Energy contribution | -31.43 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.16 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22995610 101 - 27905053 UUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCA (((((.......)))))(((..(((((........((((((((((((....(((((((...........))))))).)))))))))))))))))..))).. ( -36.90) >DroSec_CAF1 3240 92 - 1 UUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCA---------GCAACUGCA (((((.......)))))...((..((((...)))).(((((((((((....(((((((...........))))))).))))))---------))))).)). ( -31.40) >DroSim_CAF1 3250 101 - 1 UUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCA (((((.......)))))(((..(((((........((((((((((((....(((((((...........))))))).)))))))))))))))))..))).. ( -36.90) >consensus UUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCA (((((.......))))).....((..(((......((((((((((((....(((((((...........))))))).))))))))))))...)))...)). (-29.43 = -31.43 + 2.00)


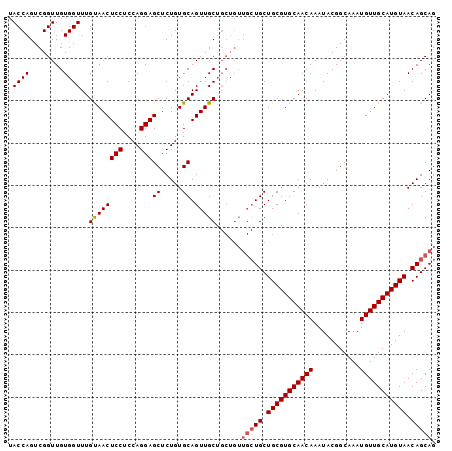
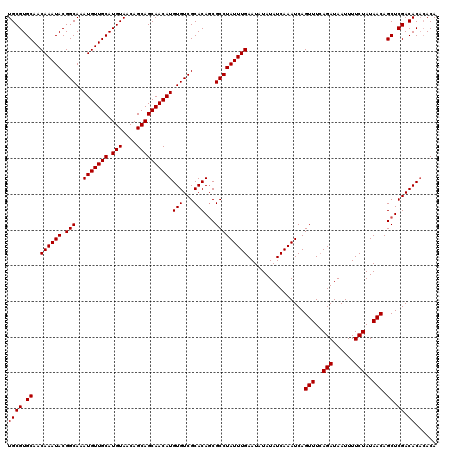
| Location | 22,995,631 – 22,995,751 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -28.85 |
| Energy contribution | -28.85 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22995631 120 + 27905053 UGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACA (((((((((((...........)))))))))))............(((((((...(((....((((((.......))))))..(((..(((......)))..))).)))))))))).... ( -30.80) >DroSec_CAF1 3252 120 + 1 UGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACA (((((((((((...........)))))))))))............(((((((...(((....((((((.......))))))..(((..(((......)))..))).)))))))))).... ( -30.80) >DroSim_CAF1 3271 120 + 1 UGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACA (((((((((((...........)))))))))))............(((((((...(((....((((((.......))))))..(((..(((......)))..))).)))))))))).... ( -30.80) >DroPer_CAF1 3017 118 + 1 UGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAUCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUAAGAUAAUUUUCUAUAACAGCUCGACAC--ACA ((((.((..((((((.(((..(((((((.(((....))))))))))(((...)))...)))))))))................(((..(((......)))..))).)).)).)).--... ( -28.10) >consensus UGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACA ((((.((..((((((.(((..(((((((.(((....))))))))))(((...)))...)))))))))................(((..(((......)))..))).)).)).))...... (-28.85 = -28.85 + -0.00)


| Location | 22,995,631 – 22,995,751 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -31.67 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
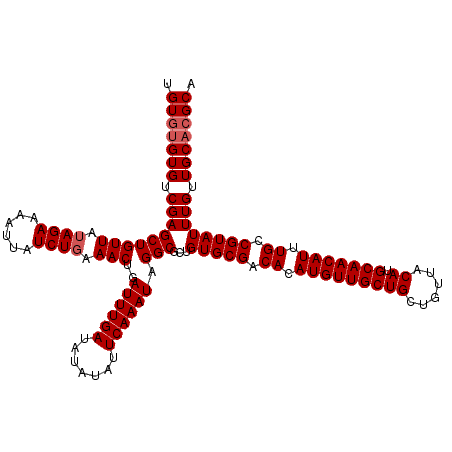
>3R_DroMel_CAF1 22995631 120 - 27905053 UGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCA .(((((((((((((.(((.((((......)))).))).).))))))))))))..........((.((((((((.(((..((...((((......))))...))..))).)))))))))). ( -34.10) >DroSec_CAF1 3252 120 - 1 UGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCA .(((((((((((((.(((.((((......)))).))).).))))))))))))..........((.((((((((.(((..((...((((......))))...))..))).)))))))))). ( -34.10) >DroSim_CAF1 3271 120 - 1 UGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCA .(((((((((((((.(((.((((......)))).))).).))))))))))))..........((.((((((((.(((..((...((((......))))...))..))).)))))))))). ( -34.10) >DroPer_CAF1 3017 118 - 1 UGU--GUGUCGAGCUGUUAUAGAAAAUUAUCUUAAACUGAUUUGAUAUAUAUUCAAAUAGGCGAUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCA .((--(((.(((.....((((..((((((........))))))..))))....(((((((((((((((((((....)))))(((........)))))).))))).)))))))))))))). ( -31.70) >consensus UGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCA .(((((((.(((((((((.((((......)))).)))..((((((.......)))))).)))...(((((.((.(((((((((......)).))))))).)).)))))))).))))))). (-31.67 = -32.42 + 0.75)


| Location | 22,995,671 – 22,995,791 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
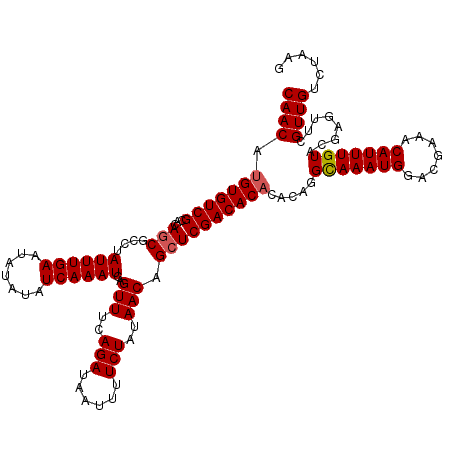
>3R_DroMel_CAF1 22995671 120 + 27905053 CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACAGGCAAAUGGACGAAACAUUUGUACGAGUUCGUUGUCUAAG ((((.(((((((...(((....((((((.......))))))..(((..(((......)))..))).)))))))))).....(((((((.......)))))))........))))...... ( -27.60) >DroSec_CAF1 3292 120 + 1 CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACAGGUAAAUGGACGAAACAUUUGUACGAGUUCGUUGUCUAAG .....(((((((...(((....((((((.......))))))..(((..(((......)))..))).))))))))))..........(((((((...((((....))))...))))))).. ( -25.30) >DroSim_CAF1 3311 120 + 1 CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACAGGCAAAUGGACGAAACAUUUGUACGAGUUCGUUGUCUAAG ((((.(((((((...(((....((((((.......))))))..(((..(((......)))..))).)))))))))).....(((((((.......)))))))........))))...... ( -27.60) >DroPer_CAF1 3057 118 + 1 CAACAUGUGUCGCACAUCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUAAGAUAAUUUUCUAUAACAGCUCGACAC--ACAGGCAAAUGGACAAAACAUUUGUACGAGUUUGUUGCCAAAG ((((((((((((..(.......((((((.......))))))..(((..(((......)))..))).)..))))))--)...(((((((.......))))))).......)))))...... ( -24.00) >consensus CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAUCAGUUUCAGAUAAUUUUCUAUAACAGCUCGACACACACAGGCAAAUGGACGAAACAUUUGUACGAGUUCGUUGUCUAAG ((((.(((((((...(((....((((((.......))))))..(((..(((......)))..))).)))))))))).....(((((((.......)))))))........))))...... (-23.59 = -23.90 + 0.31)

| Location | 22,995,671 – 22,995,791 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.43 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
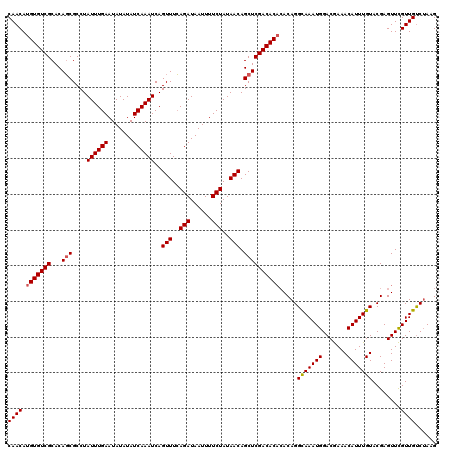
>3R_DroMel_CAF1 22995671 120 - 27905053 CUUAGACAACGAACUCGUACAAAUGUUUCGUCCAUUUGCCUGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG ......(((((...(((((((.......((((.(((((...(((((((((((((.(((.((((......)))).))).).)))))))))))).))))).)))).)))))))....))))) ( -31.60) >DroSec_CAF1 3292 120 - 1 CUUAGACAACGAACUCGUACAAAUGUUUCGUCCAUUUACCUGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG ......(((((...(((((((.......((((.((((....(((((((((((((.(((.((((......)))).))).).))))))))))))..)))).)))).)))))))....))))) ( -28.50) >DroSim_CAF1 3311 120 - 1 CUUAGACAACGAACUCGUACAAAUGUUUCGUCCAUUUGCCUGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG ......(((((...(((((((.......((((.(((((...(((((((((((((.(((.((((......)))).))).).)))))))))))).))))).)))).)))))))....))))) ( -31.60) >DroPer_CAF1 3057 118 - 1 CUUUGGCAACAAACUCGUACAAAUGUUUUGUCCAUUUGCCUGU--GUGUCGAGCUGUUAUAGAAAAUUAUCUUAAACUGAUUUGAUAUAUAUUCAAAUAGGCGAUGUGCGACACAUGUUG .((((....)))).(((((((......(((((.(((((..(((--(((((((((.(((..(((......)))..))).).)))))))))))..))))).))))))))))))......... ( -30.40) >consensus CUUAGACAACGAACUCGUACAAAUGUUUCGUCCAUUUGCCUGUGUGUGUCGAGCUGUUAUAGAAAAUUAUCUGAAACUGAUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG ......(((((...(((((((.......((((.(((((...(((((((((((((.(((.((((......)))).))).).)))))))))))).))))).)))).)))))))....))))) (-28.80 = -29.43 + 0.63)


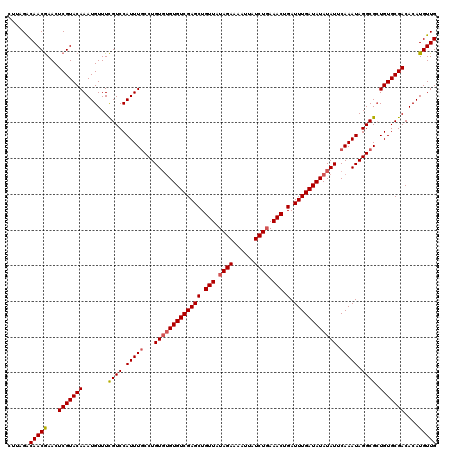
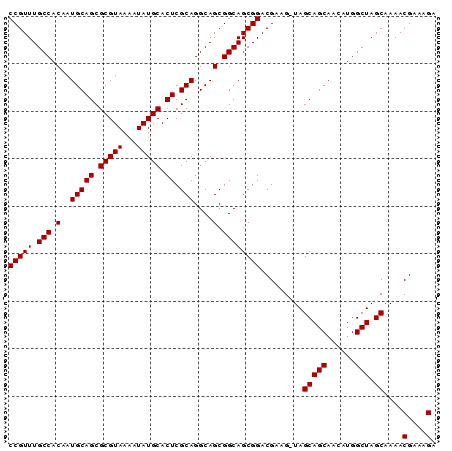
| Location | 22,995,791 – 22,995,881 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 98.51 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22995791 90 - 27905053 CCGUUUGCCACAAUGCAGCGCGUAAAAUAUGCACUCGCAGGCAGCGGCAGCGGACGGAGCUAGCAGCAACAUGGCUAGCAAAACGAAAGA ((((((((.....(((((.(((((...))))).)).))).((....)).)))))))).((((((.........))))))....(....). ( -31.50) >DroSec_CAF1 3412 89 - 1 CCGUUUGCCACAAUGCAGCGCGUAAAAUAUGCACUCGCAGGCAGCGGCAGCGGACGAAG-UAGCAGCAACAUGGCUAGCAAAACGAAAGA (((((.(((.(..(((((.(((((...))))).)).)))....).))))))))......-..(((((......))).))....(....). ( -26.60) >DroSim_CAF1 3431 89 - 1 CCGUUUGCCACAAUGCAGCGCGUAAAAUAUGCACUCGCAGGCAGCGGCAGCGGACGAAG-UAGCAGCAACAUGGCUAGCAAAACGAAAGA (((((.(((.(..(((((.(((((...))))).)).)))....).))))))))......-..(((((......))).))....(....). ( -26.60) >consensus CCGUUUGCCACAAUGCAGCGCGUAAAAUAUGCACUCGCAGGCAGCGGCAGCGGACGAAG_UAGCAGCAACAUGGCUAGCAAAACGAAAGA (((((.(((.(..(((((.(((((...))))).)).)))....).)))))))).........(((((......))).))....(....). (-26.60 = -26.60 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:16 2006