
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,994,648 – 22,994,981 |
| Length | 333 |
| Max. P | 0.999620 |

| Location | 22,994,648 – 22,994,765 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.29 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994648 117 + 27905053 UACGUCUCGAUGCCUAAUCUGCUGCCCAGUGGGUGGCCCUUCAGCAACGGCCAUUGACACUA---CUAAAUGGCCCGGCUCCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUU ...((.((((((((....((((..(((...)))..)).....))....)).)))))).))..---....((((((.(((...((......)).(((......)))))).))))))..... ( -34.60) >DroSec_CAF1 2298 117 + 1 UACGUCUCGAUGCCUAAUCUGCUGCCCAGUGGGUGGCCCUUCAGCAUCGGCCAUUGACACUA---CUAAAUGGCCCGGCUUCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUU ...((((((((((.......((..(((...)))..))......))))))).....)))....---....((((((.(((...((......)).(((......)))))).))))))..... ( -37.82) >DroSim_CAF1 2318 117 + 1 UACGUCUCGAUGCCUAAUCUGCUGCCCAGUGGGUGGCCCUUCAGCAUCGGCCAUUGACACUA---CUAAAUGGCCCGGCUCCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUU ...((((((((((.......((..(((...)))..))......))))))).....)))....---....((((((.(((...((......)).(((......)))))).))))))..... ( -37.82) >DroPer_CAF1 2161 109 + 1 AGUGGAGAGUGGCCCAAUCU--UUCGCAGUGGGUGGCC-------AUCGGCCAUUGACAUAAUUGCCAGAU--CUCUGCUCGGCUCAUUUGGCCAAUCUAAUUCAGUUUGGCCAUCAAUA ((..((((.((((.(.....--...)..((.(((((((-------...))))))).))......))))..)--)))..)).........(((((((.((.....)).)))))))...... ( -36.30) >consensus UACGUCUCGAUGCCUAAUCUGCUGCCCAGUGGGUGGCCCUUCAGCAUCGGCCAUUGACACUA___CUAAAUGGCCCGGCUCCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUU ............................((.(((((((..........))))))).))...........((((((.((((........((....))........)))).))))))..... (-23.10 = -23.29 + 0.19)


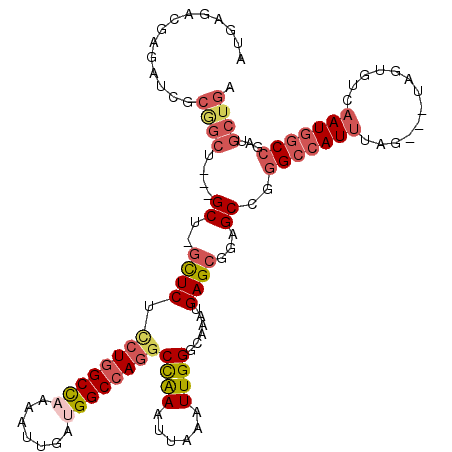
| Location | 22,994,688 – 22,994,800 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -24.41 |
| Energy contribution | -25.97 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994688 112 + 27905053 UCAGCAACGGCCAUUGACACUA---CUAAAUGGCCCGGCUCCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUUUGGGCCAGGAGAACUUGC---GGCUGC--UCUCGUCUCAU ........(((((((.......---...))))))).(((.((((........((((......))))(((((((.........))))))).......))---))..))--).......... ( -36.20) >DroSec_CAF1 2338 113 + 1 UCAGCAUCGGCCAUUGACACUA---CUAAAUGGCCCGGCUUCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUUUUGGCCAGAAGAGC-AGC---AGCCGCGAUCUCGUCUCAU ...((...(((((((.......---...))))))).((((.(((((......((((......)))).(((((((.......)))))))..))))-.).---))))))............. ( -41.90) >DroSim_CAF1 2358 113 + 1 UCAGCAUCGGCCAUUGACACUA---CUAAAUGGCCCGGCUCCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUUUUGGCCAGGAGAGC-AGC---AGCCGCGAUCUCGUCUCAU ...((...(((((((.......---...))))))).(((((.((((......((((......))))((((((((.......)))))))).))))-.).---))))))............. ( -45.80) >DroPer_CAF1 2197 110 + 1 -----AUCGGCCAUUGACAUAAUUGCCAGAU--CUCUGCUCGGCUCAUUUGGCCAAUCUAAUUCAGUUUGGCCAUCAAUA--AGCGAGUGGAGU-GGCCCGAGCGAAGAUCCCAGCUCAU -----...((((((((.((....)).)))..--(((..((((.((.((((((((((.((.....)).)))))))..))).--))))))..))))-)))).((((..........)))).. ( -40.60) >consensus UCAGCAUCGGCCAUUGACACUA___CUAAAUGGCCCGGCUCCGCUCAUUUGCCCAAUUUAAUUUGGCCUGGCCAUCAAUUUUGGCCAGGAGAGC_AGC___AGCCGCGAUCUCGUCUCAU ........(((((((.............)))))))(((((..((((......((((......))))((((((((.......)))))))).)))).......))))).............. (-24.41 = -25.97 + 1.56)


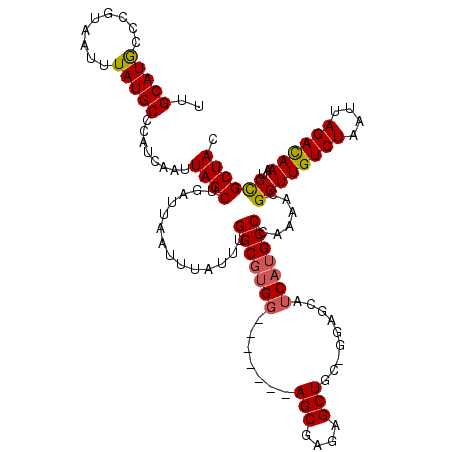
| Location | 22,994,688 – 22,994,800 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -21.47 |
| Energy contribution | -23.97 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994688 112 - 27905053 AUGAGACGAGA--GCAGCC---GCAAGUUCUCCUGGCCCAAAUUGAUGGCCAGGCCAAAUUAAAUUGGGCAAAUGAGCGGAGCCGGGCCAUUUAG---UAGUGUCAAUGGCCGUUGCUGA ..........(--(((((.---((..((((.....((((((.((((((((...))))..)))).))))))....))))...))..(((((((...---.......))))))))))))).. ( -38.70) >DroSec_CAF1 2338 113 - 1 AUGAGACGAGAUCGCGGCU---GCU-GCUCUUCUGGCCAAAAUUGAUGGCCAGGCCAAAUUAAAUUGGGCAAAUGAGCGAAGCCGGGCCAUUUAG---UAGUGUCAAUGGCCGAUGCUGA ........(((((.(((((---..(-((((..(((((((.......)))))))(((...........)))....))))).)))))(((((((...---.......)))))))))).)).. ( -43.40) >DroSim_CAF1 2358 113 - 1 AUGAGACGAGAUCGCGGCU---GCU-GCUCUCCUGGCCAAAAUUGAUGGCCAGGCCAAAUUAAAUUGGGCAAAUGAGCGGAGCCGGGCCAUUUAG---UAGUGUCAAUGGCCGAUGCUGA ........(((((.(((((---.((-((((.((((((((.......))))))))((((......))))......)))))))))))(((((((...---.......)))))))))).)).. ( -48.10) >DroPer_CAF1 2197 110 - 1 AUGAGCUGGGAUCUUCGCUCGGGCC-ACUCCACUCGCU--UAUUGAUGGCCAAACUGAAUUAGAUUGGCCAAAUGAGCCGAGCAGAG--AUCUGGCAAUUAUGUCAAUGGCCGAU----- .(((((..(....)..)))))((((-((((..((((((--((((..(((((((.(((...))).))))))))))))).))))..)))--...(((((....))))).)))))...----- ( -43.00) >consensus AUGAGACGAGAUCGCGGCU___GCU_GCUCUCCUGGCCAAAAUUGAUGGCCAGGCCAAAUUAAAUUGGGCAAAUGAGCGGAGCCGGGCCAUUUAG___UAGUGUCAAUGGCCGAUGCUGA ..............((((....((..((((.((((((((.......))))))))((((......))))......))))...))..(((((((.............)))))))...)))). (-21.47 = -23.97 + 2.50)


| Location | 22,994,800 – 22,994,912 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -23.13 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994800 112 + 27905053 UUGCAUGCCCGUAAUUUAUGCCCAUCAAUUAGCUGAUUAAUUUAUUUGGCGUGG-------AGCGAGAGCUUC-GCAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUAC ..(((((..........((((..........((..(.........)..))((((-------(((....)))))-)).)))))))))......(((((((((....))))))..))).... ( -31.40) >DroSec_CAF1 2451 112 + 1 UUGCAUGCCCGUAAUUUAUGCCCAUCAAUUAGCUGAUUAAUUUAUUUGGCGUGG-------AGCGAGAGCUGC-GGAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUAC .((.(((((((((.((((((((.((.((((((....)))))).))..)))))))-------)((....)))))-)).)))))).........(((((((((....))))))..))).... ( -32.10) >DroSim_CAF1 2471 112 + 1 UUGCAUGCCCGUAAUUUAUGCCCAUCAAUUAGCUGAUUAAUUUAUUUGGCGUGG-------AGCGAGAGCUGC-GGAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUAC .((.(((((((((.((((((((.((.((((((....)))))).))..)))))))-------)((....)))))-)).)))))).........(((((((((....))))))..))).... ( -32.10) >DroPer_CAF1 2307 117 + 1 UUGCAUACACGUAAUUUAUGCUCAUCAAUUAGCUGAUUAAUUUAUUUGGCGUGAGACAUUAAGCG-GAGCUCCCCGAACGGCCGGCCCA--ACGCUUGUCUAAUUAGAUAAAUGUGCUAC ..((((((((((...........((((......))))...........)))))(((((...((((-..(((..((....))..)))...--.)))))))))...........)))))... ( -27.15) >consensus UUGCAUGCCCGUAAUUUAUGCCCAUCAAUUAGCUGAUUAAUUUAUUUGGCGUGG_______AGCGAGAGCUGC_GGAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUAC ..(((((.........)))))........((((..............(((((((.......(((....))).........)))))))......((((((((....))))))..)))))). (-23.13 = -23.32 + 0.19)


| Location | 22,994,840 – 22,994,952 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.07 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994840 112 + 27905053 UUUAUUUGGCGUGG-------AGCGAGAGCUUC-GCAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCC ....(((((.((((-------(((....)))))-)).((.....)))))))..((((((((....)))))).((((((.....(((((((....))))))).....)))))).....)). ( -38.20) >DroSec_CAF1 2491 112 + 1 UUUAUUUGGCGUGG-------AGCGAGAGCUGC-GGAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCC ........(((((.-------(((....))).)-((.((.....))))...))))((((((....)))))).((((((.....(((((((....))))))).....))))))........ ( -36.10) >DroSim_CAF1 2511 112 + 1 UUUAUUUGGCGUGG-------AGCGAGAGCUGC-GGAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCC ........(((((.-------(((....))).)-((.((.....))))...))))((((((....)))))).((((((.....(((((((....))))))).....))))))........ ( -36.10) >DroPer_CAF1 2347 117 + 1 UUUAUUUGGCGUGAGACAUUAAGCG-GAGCUCCCCGAACGGCCGGCCCA--ACGCUUGUCUAAUUAGAUAAAUGUGCUACAUUAAUGUCGCCUCUGACAUUUACAAAGCGUGGUUCUGCC ......((.(....).))....(((-((((..((((......)))....--(((.((((((....)))))).)))(((.....(((((((....))))))).....))))..))))))). ( -29.00) >consensus UUUAUUUGGCGUGG_______AGCGAGAGCUGC_GGAGCAUCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCC .......(((((((.......(((....))).........)))))))......((((((((....)))))).((((((.....(((((((....))))))).....)))))).....)). (-29.88 = -30.07 + 0.19)



| Location | 22,994,840 – 22,994,952 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -29.80 |
| Energy contribution | -30.36 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994840 112 - 27905053 GGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGAUGCUGC-GAAGCUCUCGCU-------CCACGCCAAAUAAA ((((((((.(((((.(((.((((((......)))))).))).))))).(((((((....))))))).)))))((((..((.(((..-..)))))..)).-------))..)))....... ( -39.20) >DroSec_CAF1 2491 112 - 1 GGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGAUGCUCC-GCAGCUCUCGCU-------CCACGCCAAAUAAA ((((((((.(((((.(((.((((((......)))))).))).))))).(((((((....))))))).)))))((((..((.(((..-..)))))..)).-------))..)))....... ( -37.90) >DroSim_CAF1 2511 112 - 1 GGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGAUGCUCC-GCAGCUCUCGCU-------CCACGCCAAAUAAA ((((((((.(((((.(((.((((((......)))))).))).))))).(((((((....))))))).)))))((((..((.(((..-..)))))..)).-------))..)))....... ( -37.90) >DroPer_CAF1 2347 117 - 1 GGCAGAACCACGCUUUGUAAAUGUCAGAGGCGACAUUAAUGUAGCACAUUUAUCUAAUUAGACAAGCGU--UGGGCCGGCCGUUCGGGGAGCUC-CGCUUAAUGUCUCACGCCAAAUAAA (((........(((.(((.((((((......)))))).))).)))..............(((((((((.--.((((...((......)).))))-))))...)))))...)))....... ( -30.20) >consensus GGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGAUGCUCC_GCAGCUCUCGCU_______CCACGCCAAAUAAA (((......(((((.(((.((((((......)))))).))).)))))..((((((....))))))(((....((((..............)))).)))............)))....... (-29.80 = -30.36 + 0.56)

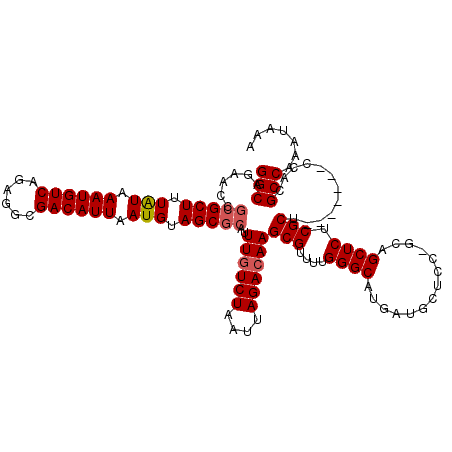

| Location | 22,994,872 – 22,994,981 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994872 109 + 27905053 UCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCCGCAGUUGGGU----GAUUUUUCACUGGUUUU-------CU ....((((((...((((((((....))))))..(((((.....(((((((....))))))).....)))))((.....))))..))))))----.................-------.. ( -31.90) >DroSec_CAF1 2523 109 + 1 UCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCCGCAGUUGGGU----GAUUUUUCACUGGUUUU-------CU ....((((((...((((((((....))))))..(((((.....(((((((....))))))).....)))))((.....))))..))))))----.................-------.. ( -31.90) >DroSim_CAF1 2543 109 + 1 UCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCCGCAGUUGGGU----GAUUUUUCACUGGUUUU-------CU ....((((((...((((((((....))))))..(((((.....(((((((....))))))).....)))))((.....))))..))))))----.................-------.. ( -31.90) >DroPer_CAF1 2386 118 + 1 GCCGGCCCA--ACGCUUGUCUAAUUAGAUAAAUGUGCUACAUUAAUGUCGCCUCUGACAUUUACAAAGCGUGGUUCUGCCGCAGCUCAGACACGGGUUUUUCAUUGUUUUUGGCCAUUAG ...((((..--(((.((((((....)))))).)))(((.....(((((((....))))))).....)))((((.....))))(((((......))))).............))))..... ( -29.70) >consensus UCAUGCCCAAAACGCUUGUCUAAUUAGACAAAUGCGCUACAUUAAUGUCGCCUCUGACAUUUAUAAAGCGCGGUUCUGCCGCAGUUGGGU____GAUUUUUCACUGGUUUU_______CU ....((((((...((((((((....))))))..))(((.....(((((((....))))))).....)))((((.....))))..)))))).............................. (-25.31 = -25.75 + 0.44)



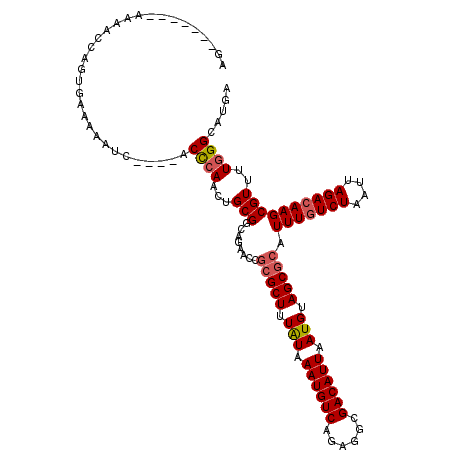
| Location | 22,994,872 – 22,994,981 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22994872 109 - 27905053 AG-------AAAACCAGUGAAAAAUC----ACCCAACUGCGGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGA ..-------...............((----((((((..(((........(((((.(((.((((((......)))))).))).))))).(((((((....)))))))))).)))))..))) ( -31.90) >DroSec_CAF1 2523 109 - 1 AG-------AAAACCAGUGAAAAAUC----ACCCAACUGCGGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGA ..-------...............((----((((((..(((........(((((.(((.((((((......)))))).))).))))).(((((((....)))))))))).)))))..))) ( -31.90) >DroSim_CAF1 2543 109 - 1 AG-------AAAACCAGUGAAAAAUC----ACCCAACUGCGGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGA ..-------...............((----((((((..(((........(((((.(((.((((((......)))))).))).))))).(((((((....)))))))))).)))))..))) ( -31.90) >DroPer_CAF1 2386 118 - 1 CUAAUGGCCAAAAACAAUGAAAAACCCGUGUCUGAGCUGCGGCAGAACCACGCUUUGUAAAUGUCAGAGGCGACAUUAAUGUAGCACAUUUAUCUAAUUAGACAAGCGU--UGGGCCGGC ....(((((...(((.(((.......)))(((((((((((((((((.......))))).((((((......))))))..))))))............))))))....))--).))))).. ( -28.70) >consensus AG_______AAAACCAGUGAAAAAUC____ACCCAACUGCGGCAGAACCGCGCUUUAUAAAUGUCAGAGGCGACAUUAAUGUAGCGCAUUUGUCUAAUUAGACAAGCGUUUUGGGCAUGA ...............................((((...(((........(((((.(((.((((((......)))))).))).))))).(((((((....))))))))))..))))..... (-25.10 = -25.72 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:07 2006