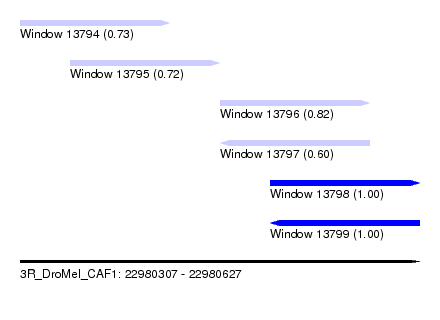
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,980,307 – 22,980,627 |
| Length | 320 |
| Max. P | 0.998882 |

| Location | 22,980,307 – 22,980,427 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -35.78 |
| Energy contribution | -35.38 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22980307 120 + 27905053 ACAAGUAUCUCUAUCUGAGCGAUCUGUACGAUCGCAAUGAGCGUCUGUUCUUCCGCUUCCUAUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCU .(((..(((...((((..((((((.....))))))......((..((((((.(((........))))))))).)).))))..)))(((((((((..........)))))))))))).... ( -36.20) >DroSec_CAF1 11218 120 + 1 ACAAGUAUCUCUAUCUGAGCGAUCUGUACGAUCGCAAUGAGCGUCUUUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCU .(((((.((.(..((.(.(((...(((((....((((((.(((((...(((((...((((....))))......)))))...))))).)))))))))))))))))..).))))))).... ( -35.80) >DroSim_CAF1 4357 120 + 1 ACAAGUAUCUGUAUCUGAGCGAUCUGUACGAUCGCAAUGAGCGUCUGUUCUUCCGAUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCU .(((((....(((((.((((((((.....))))))......(.((((((((.(((((....)))))))))))..)).).)).)))))(((((((..........)))))))))))).... ( -41.70) >DroEre_CAF1 5149 120 + 1 ACAAGUACCUCUACUUGAGCGAUCUGUACGAUCGCAACGAGCGUCUGUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCCACAGUGGGCUUGGCGU .((((((....)))))).((((((.....)))))).....(((((((((((((.((.....)).)))))))).............((((((((((........))))))))))..))))) ( -47.10) >DroYak_CAF1 11787 120 + 1 ACAAGUACCUCUACCUGAGCGAUCUGUACGAUCGCAAUGAGCGUCUGUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCUAUUGUGUACACACCCACAGUGGGUUUGGCCU .......((((.......((((((.....))))))..........((((((((.((.....)).))))))))..))))..(..(.((((((((((........)))))))))))..)... ( -39.70) >consensus ACAAGUAUCUCUAUCUGAGCGAUCUGUACGAUCGCAAUGAGCGUCUGUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCU .......((((.......((((((.....))))))..........((((((.(((........)))))))))..))))..(..(.(((((((((..........))))))))))..)... (-35.78 = -35.38 + -0.40)



| Location | 22,980,347 – 22,980,467 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -43.32 |
| Consensus MFE | -38.98 |
| Energy contribution | -38.58 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22980347 120 + 27905053 GCGUCUGUUCUUCCGCUUCCUAUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCUGCCAGCGCUUUGGCCUGAUCUACCGACGUCCUCAUGGUCU (((((((((((.(((........)))))))))....(((((....(((((((((..........)))))))))..((((.((....))...)))).)))))...)))))((....))... ( -42.20) >DroSec_CAF1 11258 120 + 1 GCGUCUUUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCUGCCAGCGCUUUGGCCUGAUCUACAGACGUCCUCAUGGUCU .......((((((.((.....)).))))))(((.(((((((((..(((((((((..........)))))))))..((((.((....))...)))).......))))..)))))))).... ( -41.10) >DroSim_CAF1 4397 120 + 1 GCGUCUGUUCUUCCGAUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCUGCCAGCGCUUUGGCCUGAUCUACAGACGUCCUCAUGGUCU (((((((((((.(((((....))))))))))...(.(((((....(((((((((..........)))))))))..((((.((....))...)))).))))).)))))))((....))... ( -47.20) >DroEre_CAF1 5189 120 + 1 GCGUCUGUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCCACAGUGGGCUUGGCGUGCCAGCGAUUUGGCCUGAUCUACAGACGUCCUCAUGGUCU .((..((((((((.((.....)).)))))))).)).((((((((.((((((((((........))))))))))..((((((((((....)))))(((.....)))))))).))).))))) ( -45.50) >DroYak_CAF1 11827 120 + 1 GCGUCUGUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCUAUUGUGUACACACCCACAGUGGGUUUGGCCUGCCAGCGUUUCGGUCUGAUCUACAGGCGUCCUCAUGGUCU (((((((((((((.((.....)).)))))))...(.(((((.(((((((((((((........))))))))))((((....)))).......))).))))).)))))))((....))... ( -40.60) >consensus GCGUCUGUUCUUCCGCUUCCUGUCGGAGAACAUCGAGGAUCUGAUGCCCAUUGUGUACACGCCGACGGUGGGCUUGGCCUGCCAGCGCUUUGGCCUGAUCUACAGACGUCCUCAUGGUCU (((((((((((.(((........)))))))))..(.(((((....(((((((((..........)))))))))..((((.((....))...)))).))))).).)))))((....))... (-38.98 = -38.58 + -0.40)



| Location | 22,980,467 – 22,980,587 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -40.34 |
| Energy contribution | -39.94 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22980467 120 + 27905053 GUUCAUCACCUACAACGAUCGAGGACACAUCUUUGAUGUGAUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCCUGGGACUGGGAGAUUUGGG .((((((((........((((((((....)))))))))))))))).......((((..(((.((((((..((((((((.....))))))))..))))))..)))..)))).......... ( -43.40) >DroSec_CAF1 11378 120 + 1 GUUCAUCACCUACAACGAUCGCGGACACAUCUUUGAUGUGAUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGCGAGCGCAUCCUGGGACUGGGAGAUUUGGG ((((.(((.......((....))..((((((...)))))).))).))))...((((..(((.((((((.(((((((((.....))))))))).))))))..)))..)))).......... ( -43.10) >DroSim_CAF1 4517 120 + 1 GUUUAUCACCUACAACGAUCGCGGACACAUCUUUGAUGUGAUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGCGAGCGCAUCCUGGGACUGGGAGAUUUGGG ........((((....((((.....((((((...))))))............((((..(((.((((((.(((((((((.....))))))))).))))))..)))..))))..)))))))) ( -41.80) >DroEre_CAF1 5309 120 + 1 GUUCAUCACGUACAACGAUCGCGGACACAUCUUUGAUGUGAUGAAGAAUUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCCUGGGACUGGGAGAUUUGGG .((((((((((.(((.(((.(......)))).)))))))))))))(((((..((((..(((.((((((..((((((((.....))))))))..))))))..)))..))))..)))))... ( -46.50) >DroYak_CAF1 11947 120 + 1 GUUCAUCACGUACAACGAUCGCGGACACAUCUUUGAUGUGAUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCUUGGGACUGGGAGAUUUGGG .((((((((((.(((.(((.(......)))).))))))))))))).......((((..((((((((((..((((((((.....))))))))..)))))).))))..)))).......... ( -45.20) >consensus GUUCAUCACCUACAACGAUCGCGGACACAUCUUUGAUGUGAUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCCUGGGACUGGGAGAUUUGGG .((((((((...(((.(((.(......)))).)))..)))))))).......((((..(((.((((((.(((((((((.....))))))))).))))))..)))..)))).......... (-40.34 = -39.94 + -0.40)


| Location | 22,980,467 – 22,980,587 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -30.54 |
| Energy contribution | -31.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22980467 120 - 27905053 CCCAAAUCUCCCAGUCCCAGGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAUCACAUCAAAGAUGUGUCCUCGAUCGUUGUAGGUGAUGAAC ...................(((((.((...(((((.(.....).)))))...)).)))))(((....))).....((((((((..(((.(((.((....))))).)))...)))))))). ( -32.40) >DroSec_CAF1 11378 120 - 1 CCCAAAUCUCCCAGUCCCAGGAUGCGCUCGCCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAUCACAUCAAAGAUGUGUCCGCGAUCGUUGUAGGUGAUGAAC ...................(((((.((..((((((.(.....).))))))..)).)))))(((....))).....((((((((..(((.(((((....)).))).)))...)))))))). ( -37.70) >DroSim_CAF1 4517 120 - 1 CCCAAAUCUCCCAGUCCCAGGAUGCGCUCGCCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAUCACAUCAAAGAUGUGUCCGCGAUCGUUGUAGGUGAUAAAC .............(((((.(((((.((..((((((.(.....).))))))..)).)))))(((....))).(.((.....((((((...)))))).....)).)......)).))).... ( -32.80) >DroEre_CAF1 5309 120 - 1 CCCAAAUCUCCCAGUCCCAGGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAAUUCUUCAUCACAUCAAAGAUGUGUCCGCGAUCGUUGUACGUGAUGAAC ...................(((((.((...(((((.(.....).)))))...)).)))))(((....))).....((((((((..(((.(((((....)).))).)))...)))))))). ( -32.90) >DroYak_CAF1 11947 120 - 1 CCCAAAUCUCCCAGUCCCAAGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAUCACAUCAAAGAUGUGUCCGCGAUCGUUGUACGUGAUGAAC ....................((((.((...(((((.(.....).)))))...)).)))).(((....))).....((((((((..(((.(((((....)).))).)))...)))))))). ( -29.40) >consensus CCCAAAUCUCCCAGUCCCAGGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAUCACAUCAAAGAUGUGUCCGCGAUCGUUGUAGGUGAUGAAC ...................(((((.((...(((((.(.....).)))))...)).)))))(((....))).....((((((((..(((.(((((....)).))).)))...)))))))). (-30.54 = -31.14 + 0.60)



| Location | 22,980,507 – 22,980,627 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -54.40 |
| Consensus MFE | -53.12 |
| Energy contribution | -52.76 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22980507 120 + 27905053 AUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCCUGGGACUGGGAGAUUUGGGCGCCUGCGGCAUGGGCAUUCCCGUGGGCAAGCUGGCCUUG ..........((((((.(((.(((((((..((((((((.....))))))))..)))))((((......))))...)).)))(((..(((...........)))..)))...))))))... ( -53.90) >DroSec_CAF1 11418 120 + 1 AUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGCGAGCGCAUCCUGGGACUGGGAGAUUUGGGCGCCUGCGGCAUGGGCAUUCCCGUGGGCAAGCUGGCCUUG ..........((((((.(((.(((((((.(((((((((.....))))))))).)))))((((......))))...)).)))(((..(((...........)))..)))...))))))... ( -56.40) >DroSim_CAF1 4557 120 + 1 AUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGCGAGCGCAUCCUGGGACUGGGAGAUUUGGGAGCCUGCGGCAUGGGCAUUCCCGUGGGCAAGCUGGCCUUG ..........((((((..((((((((((.(((((((((.....))))))))).)))).((((......)))).))))))..(((..(((...........)))..)))...))))))... ( -54.80) >DroEre_CAF1 5349 120 + 1 AUGAAGAAUUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCCUGGGACUGGGAGAUUUGGGCGCCUGCGGCAUGGGCAUUCCCGUGGGCAAGCUGGCCUUG ..........((((((.(((.(((((((..((((((((.....))))))))..)))))((((......))))...)).)))(((..(((...........)))..)))...))))))... ( -53.90) >DroYak_CAF1 11987 120 + 1 AUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCUUGGGACUGGGAGAUUUGGGCGCUUGCGGCAUGGGCAUUCCCGUGGGCAAGCUGGCCUUG ..........((((((.(((((((((.(((((((((((.....))))).(..((((((((((........)))))....)))))..))))))).)))))).....)))...))))))... ( -53.00) >consensus AUGAAGAACUGGCCGGAGCCGAAUGUGCGUGCCAUCUGUGUGACGGAUGGUGAGCGCAUCCUGGGACUGGGAGAUUUGGGCGCCUGCGGCAUGGGCAUUCCCGUGGGCAAGCUGGCCUUG ..........((((((.(((.(((((((.(((((((((.....))))))))).)))))((((......))))...)).)))(((..(((...........)))..)))...))))))... (-53.12 = -52.76 + -0.36)



| Location | 22,980,507 – 22,980,627 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -38.52 |
| Energy contribution | -38.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22980507 120 - 27905053 CAAGGCCAGCUUGCCCACGGGAAUGCCCAUGCCGCAGGCGCCCAAAUCUCCCAGUCCCAGGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAU ...((((.((((((.((.(((....))).))..))))))(((.....(.....).....(((((.((...(((((.(.....).)))))...)).))))))))...)))).......... ( -39.60) >DroSec_CAF1 11418 120 - 1 CAAGGCCAGCUUGCCCACGGGAAUGCCCAUGCCGCAGGCGCCCAAAUCUCCCAGUCCCAGGAUGCGCUCGCCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAU ...((((.((((((.((.(((....))).))..))))))(((.....(.....).....(((((.((..((((((.(.....).))))))..)).))))))))...)))).......... ( -43.30) >DroSim_CAF1 4557 120 - 1 CAAGGCCAGCUUGCCCACGGGAAUGCCCAUGCCGCAGGCUCCCAAAUCUCCCAGUCCCAGGAUGCGCUCGCCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAU ...(((((((((((.((.(((....))).))..)))))))............((((...(((((.((..((((((.(.....).))))))..)).)))))))))..)))).......... ( -42.80) >DroEre_CAF1 5349 120 - 1 CAAGGCCAGCUUGCCCACGGGAAUGCCCAUGCCGCAGGCGCCCAAAUCUCCCAGUCCCAGGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAAUUCUUCAU ...((((.((((((.((.(((....))).))..))))))(((.....(.....).....(((((.((...(((((.(.....).)))))...)).))))))))...)))).......... ( -39.60) >DroYak_CAF1 11987 120 - 1 CAAGGCCAGCUUGCCCACGGGAAUGCCCAUGCCGCAAGCGCCCAAAUCUCCCAGUCCCAAGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAU ...((((.((((((.((.(((....))).))..))))))(((...((((..........))))(((....(((((.(.....).)))))..)))......)))...)))).......... ( -37.70) >consensus CAAGGCCAGCUUGCCCACGGGAAUGCCCAUGCCGCAGGCGCCCAAAUCUCCCAGUCCCAGGAUGCGCUCACCAUCCGUCACACAGAUGGCACGCACAUUCGGCUCCGGCCAGUUCUUCAU ...((((.((((((.((.(((....))).))..)))))).............((((...(((((.((...(((((.(.....).)))))...)).)))))))))..)))).......... (-38.52 = -38.56 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:50 2006