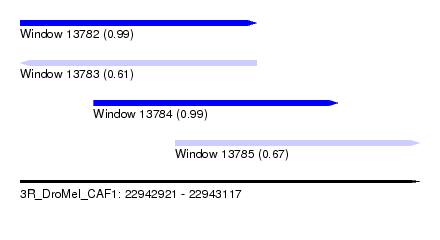
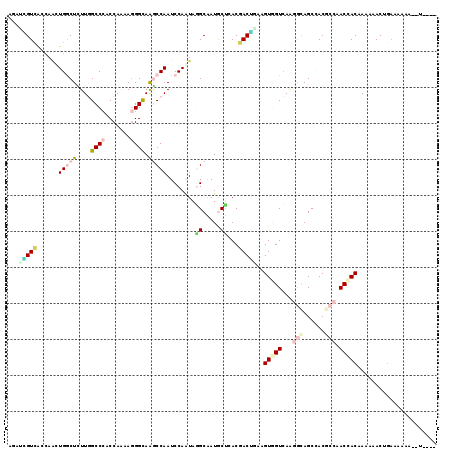
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,942,921 – 22,943,117 |
| Length | 196 |
| Max. P | 0.994888 |

| Location | 22,942,921 – 22,943,037 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.25 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.49 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
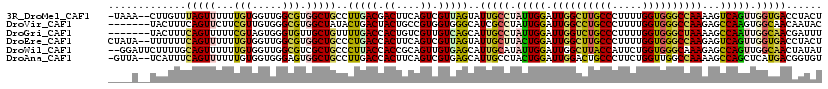
>3R_DroMel_CAF1 22942921 116 + 27905053 -UAAA--CUUGUUUAGUUUUUUGUGGUUGGCGUGGCUGCCUUGACGACUUCAGUCGUUAGUAUUGCCUAUUGGAUUGGCUUGCCCUUUUGGUGGGCCAAAAGUCAGUUGGUGACCUACU -.(((--((.....)))))...((((((((((....))))((((((((....))))))))....(((.(((((.((((((..((.....))..))))))...))))).))))))).)). ( -39.30) >DroVir_CAF1 448 112 + 1 -------UACUUUCAGUUCUUCGUUGUGGGCGUGGCUAUACUGACUACUGCCGUGGUGGGCAUCGCCUAUUGGAUUGGCCUGCCCUUUUGGUGGGCCAAGAGCCAAGUGGCAACAAUAC -------...............((((((((((((((......).))))((((......))))..)))).((((.((((((..((.....))..))))))...))))...)))))..... ( -42.10) >DroGri_CAF1 236 112 + 1 -------UACUUUCAGUUUUUCGUAGUGGGUGUUGCUGUUUUGACCACUGUCGUUGUCAGCAUUGCCUAUUGGAUUGGUCUGCCCUUUUGGUGGGCUAAAAGCCAAUUGGCAACGAUUU -------.............(((((((((((..(((((...((((....))))....)))))..)))))))...((((((..((.....))..))))))..(((....))).))))... ( -37.40) >DroEre_CAF1 2465 117 + 1 CUAUA--UUUUUUCAGUUUUUUGUGGUUGGCGUGGCUGCCCUGACCACUUCAGUCGUUAGUAUUGCUUACUGGAUUGGCUUGCCCUUUUGGUGGGCCAAGAGUCAGUUGGUGACCUACU .....--...............((((((((((....))))..))))))..........(((((..((.(((((.((((((..((.....))..))))))...))))).))..)..)))) ( -37.80) >DroWil_CAF1 129 117 + 1 --GGAUUCUUUUGCAGUUUUUUGUGGUUGGCGUCGCUGCCCUUACCACCGCAGUUGUGAGCAUUGCAUAUUGGAUUGGCUUACCAUUCUGGUGGGCAAAGAGCCAGUUGGCAACUAUAU --.(((((...((((((.....(((((.((((....))))...)))))(((....)))...))))))....))))).(((((((.....))))))).....(((....)))........ ( -36.10) >DroAna_CAF1 197 116 + 1 -GUUA--UCAUUUCAGUUUUUUGUGGUGGGAGUGGCUGCCUUGACCACUUCAGUCGUGAGCAUUGCCUACUGGAUUGGACUGCCCUUCUGGUUGGCCAAAAGCCAGCUCAUGACGGUGU -....--.(((..(((....)))..)))((((((((......).))))))).(((((((((.......((..((..((....))..))..))((((.....)))))))))))))..... ( -41.30) >consensus ____A__UUUUUUCAGUUUUUUGUGGUGGGCGUGGCUGCCCUGACCACUGCAGUCGUGAGCAUUGCCUAUUGGAUUGGCCUGCCCUUUUGGUGGGCCAAAAGCCAGUUGGCAACCAUAU .............(((((...(((.....))).)))))..(((((.((....)).)))))..(((((.(((((.((((((((((.....))))))))))...))))).)))))...... (-26.58 = -26.25 + -0.33)
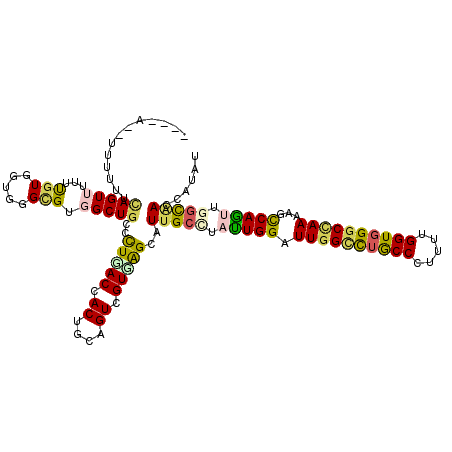
| Location | 22,942,921 – 22,943,037 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -13.11 |
| Energy contribution | -14.58 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22942921 116 - 27905053 AGUAGGUCACCAACUGACUUUUGGCCCACCAAAAGGGCAAGCCAAUCCAAUAGGCAAUACUAACGACUGAAGUCGUCAAGGCAGCCACGCCAACCACAAAAAACUAAACAAG--UUUA- .((.(((..........(((((((....)))))))(((..(((.......(((......)))(((((....)))))...))).)))..))).))......(((((.....))--))).- ( -28.40) >DroVir_CAF1 448 112 - 1 GUAUUGUUGCCACUUGGCUCUUGGCCCACCAAAAGGGCAGGCCAAUCCAAUAGGCGAUGCCCACCACGGCAGUAGUCAGUAUAGCCACGCCCACAACGAAGAACUGAAAGUA------- ...((((((...(.(((((.((((((..((.....))..)))))).......(((..((((......))))...))).....))))).)....)))))).............------- ( -31.50) >DroGri_CAF1 236 112 - 1 AAAUCGUUGCCAAUUGGCUUUUAGCCCACCAAAAGGGCAGACCAAUCCAAUAGGCAAUGCUGACAACGACAGUGGUCAAAACAGCAACACCCACUACGAAAAACUGAAAGUA------- ...((((((((.(((((.((((.((((.......)))).))..)).))))).)))).(((((.....(((....)))....))))).........)))).............------- ( -28.00) >DroEre_CAF1 2465 117 - 1 AGUAGGUCACCAACUGACUCUUGGCCCACCAAAAGGGCAAGCCAAUCCAGUAAGCAAUACUAACGACUGAAGUGGUCAGGGCAGCCACGCCAACCACAAAAAACUGAAAAAA--UAUAG .((.(((......(((((((((.((((.......)))).........((((.((.....))....))))))).))))))(((......))).)))))...............--..... ( -26.20) >DroWil_CAF1 129 117 - 1 AUAUAGUUGCCAACUGGCUCUUUGCCCACCAGAAUGGUAAGCCAAUCCAAUAUGCAAUGCUCACAACUGCGGUGGUAAGGGCAGCGACGCCAACCACAAAAAACUGCAAAAGAAUCC-- ...(((((......((((((.((((((((((((.(((....))).))..........(((........))).))))..)))))).)).)))).........)))))...........-- ( -28.56) >DroAna_CAF1 197 116 - 1 ACACCGUCAUGAGCUGGCUUUUGGCCAACCAGAAGGGCAGUCCAAUCCAGUAGGCAAUGCUCACGACUGAAGUGGUCAAGGCAGCCACUCCCACCACAAAAAACUGAAAUGA--UAAC- .....((((((((.(((((((((((((..(((..(((((..((.........))...)))))....)))...))))))))..))))))))......((......))..))))--)...- ( -30.90) >consensus AGAUCGUCACCAACUGGCUCUUGGCCCACCAAAAGGGCAAGCCAAUCCAAUAGGCAAUGCUCACGACUGAAGUGGUCAAGGCAGCCACGCCAACCACAAAAAACUGAAAAAA__U____ ...(((((......(((((....((((.......)))).)))))........((.....))...)))))..(((((...(((......))).)))))...................... (-13.11 = -14.58 + 1.47)


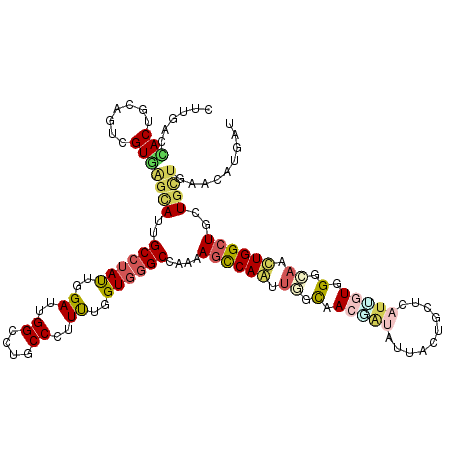
| Location | 22,942,957 – 22,943,077 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -43.75 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22942957 120 + 27905053 CUUGACGACUUCAGUCGUUAGUAUUGCCUAUUGGAUUGGCUUGCCCUUUUGGUGGGCCAAAAGUCAGUUGGUGACCUACUUCCUCCUCAUUGUGGGCAACUGGCUGCUGCUCAAUGUGGU .((((((((....))))))))....(((((((((.((((((..((.....))..)))))).((.((((..((..(((((............)))))..))..))))))..)))))).))) ( -47.10) >DroVir_CAF1 480 120 + 1 ACUGACUACUGCCGUGGUGGGCAUCGCCUAUUGGAUUGGCCUGCCCUUUUGGUGGGCCAAGAGCCAAGUGGCAACAAUACUGCUGCUUAUUGUGGGCAACUGGCUGCUGCUAAACAUUAU .((.(((((....))))).))....((((((.((.((((((..((.....))..))))))...))(((..(((.......)))..)))...))))))...((((....))))........ ( -46.10) >DroGri_CAF1 268 120 + 1 UUUGACCACUGUCGUUGUCAGCAUUGCCUAUUGGAUUGGUCUGCCCUUUUGGUGGGCUAAAAGCCAAUUGGCAACGAUUUUAUUGCUCAUCUUUGGCAAUUGGGUGCUGUUGAACAUCAU ...(((....)))(((..((((.(((((.(((((.((((((..((.....))..))))))...))))).)))))..(((..(((((.((....)))))))..)))))))...)))..... ( -40.10) >DroWil_CAF1 166 120 + 1 CCUUACCACCGCAGUUGUGAGCAUUGCAUAUUGGAUUGGCUUACCAUUCUGGUGGGCAAAGAGCCAGUUGGCAACUAUAUUUUUACUCCUGGUGGGGAACUGGCUGUUGUUAAACGUGAU .....((((((.(((.((((((..(.(.....).)...)))))).))).))))))((((..(((((((((....)..........((((....)))))))))))).)))).......... ( -40.00) >DroMoj_CAF1 24 120 + 1 CUUAACCACUGCCGUAGUGUCCAUUGCCUAUUGGAUUGGGCUGCCCUUUUGGUGGGCCAAGAGCCAAGCAGCCACUAUAGUACUGCUCAUCAUAGGCAAUUGGCUGCUGCUGAACAUAAU .............((((((((.((((((((((((.((((.(..((.....))..).))))...)))(((((..((....)).)))))....))))))))).))).))))).......... ( -42.10) >DroAna_CAF1 233 120 + 1 CUUGACCACUUCAGUCGUGAGCAUUGCCUACUGGAUUGGACUGCCCUUCUGGUUGGCCAAAAGCCAGCUCAUGACGGUGUUCCUGCUCAUUGUGGGGAACUGGCUACUUCUGAAUGUGGU ....((((((((((((((((((.......((..((..((....))..))..))((((.....))))))))))))((((.((((..(.....)..)))))))).......))))).))))) ( -47.10) >consensus CUUGACCACUGCAGUCGUGAGCAUUGCCUAUUGGAUUGGCCUGCCCUUUUGGUGGGCCAAAAGCCAAUUGGCAACGAUAUUACUGCUCAUUGUGGGCAACUGGCUGCUGCUGAACAUGAU ......(((.......)))((((..((((((..((..((....))..))..))))))....((((((.((.(.(((((..........))))).).)).))))))..))))......... (-22.98 = -23.68 + 0.70)

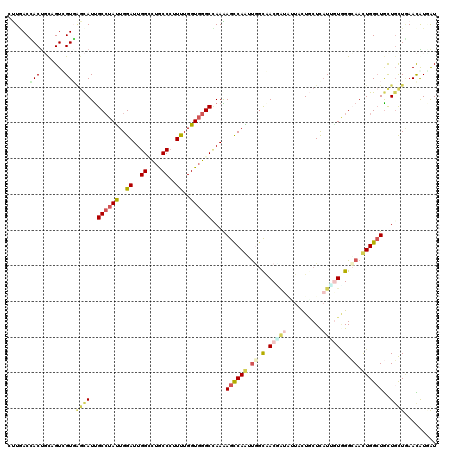
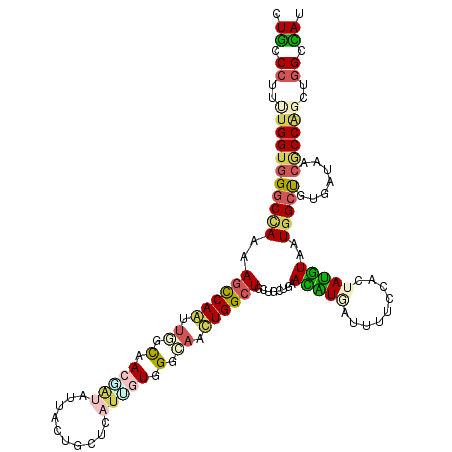
| Location | 22,942,997 – 22,943,117 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22942997 120 + 27905053 UUGCCCUUUUGGUGGGCCAAAAGUCAGUUGGUGACCUACUUCCUCCUCAUUGUGGGCAACUGGCUGCUGCUCAAUGUGGUUUUCCACUACGUUAUGGCUGUGAUCACGCCGGCUGGUCAU .((.((..(((((((((((..((.((((..((..(((((............)))))..))..))))))....((((((((.....)))))))).)))))(....).))))))..)).)). ( -39.70) >DroVir_CAF1 520 120 + 1 CUGCCCUUUUGGUGGGCCAAGAGCCAAGUGGCAACAAUACUGCUGCUUAUUGUGGGCAACUGGCUGCUGCUAAACAUUAUUUUCCAUUAUGUGAUGGCUGUGAUAACGCCCGCUGGACAG ((((((((((((....)))))))..(((..(((.......)))..)))...((((((...((((....))))...(((((...(((((....)))))..)))))...)))))).)).))) ( -40.00) >DroGri_CAF1 308 120 + 1 CUGCCCUUUUGGUGGGCUAAAAGCCAAUUGGCAACGAUUUUAUUGCUCAUCUUUGGCAAUUGGGUGCUGUUGAACAUCAUCUUCCACUAUGUGAUGGCUGUAAUAACACCACCUGGCCAU ..((((.......)))).....(((((.((((((........)))).))...)))))..(..((((.(((((.((((((((...........))))).)))..))))).))))..).... ( -35.70) >DroWil_CAF1 206 120 + 1 UUACCAUUCUGGUGGGCAAAGAGCCAGUUGGCAACUAUAUUUUUACUCCUGGUGGGGAACUGGCUGUUGUUAAACGUGAUAUUCCACUAUGUAAUGGCUGUGAUAACGCCGGCCGGUUAU ........(((((.(((....(((((((((....)..........((((....))))))))))))(((((((..(((.((((......)))).)))....)))))))))).))))).... ( -41.20) >DroMoj_CAF1 64 120 + 1 CUGCCCUUUUGGUGGGCCAAGAGCCAAGCAGCCACUAUAGUACUGCUCAUCAUAGGCAAUUGGCUGCUGCUGAACAUAAUUUUCCACUAUGUAAUGGGUGUGAUUACACCAGCUGGCCAG ..((((.......))))...(.(((((((((((((....))..((((.......))))...)))))))(((..(((((.........)))))....(((((....))))))))))))).. ( -38.00) >DroAna_CAF1 273 120 + 1 CUGCCCUUCUGGUUGGCCAAAAGCCAGCUCAUGACGGUGUUCCUGCUCAUUGUGGGGAACUGGCUACUUCUGAAUGUGGUCUUCCAUUAUAUUAUGGCCGUGAUUACCCCACCUGGGUAU ..(((.((.((((((((.....)))))).)).)).))).....((((((..((((((....((((((........))))))..((((......)))).........)))))).)))))). ( -40.50) >consensus CUGCCCUUUUGGUGGGCCAAAAGCCAAUUGGCAACGAUAUUACUGCUCAUUGUGGGCAACUGGCUGCUGCUGAACAUGAUUUUCCACUAUGUAAUGGCUGUGAUAACGCCAGCUGGCCAU .((.((..(((((((((((..((((((.((.(.(((((..........))))).).)).))))))........(((((.........)))))..))))).......))))))..)).)). (-18.98 = -19.54 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:36 2006