
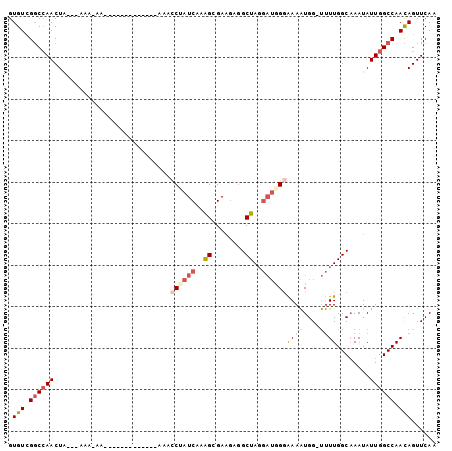
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,902,860 – 22,902,956 |
| Length | 96 |
| Max. P | 0.965417 |

| Location | 22,902,860 – 22,902,956 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.19 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -11.72 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22902860 96 + 27905053 GUGUCGGCCAACUA---AAAAAAAAA---AAAAACAAAACCUAUCAAAGCGAAGAGGCUAGGAUGGGAAAAUGG-UUUUGGCAAAUAUUGGCCAACAGUUCAA .(((.((((((...---.........---..........((((((..(((......)))..))))))......(-(....)).....)))))).)))...... ( -23.80) >DroSec_CAF1 19863 82 + 1 GUGUCGGCCAACUA---AAA-----------------AACCUAUCAAAGCGAAGAGGCUAGGAUGGGAAAAUGG-UUUUGGCAAAUAUUGGCCAACAGUUCAA .(((.((((((...---...-----------------..((((((..(((......)))..))))))......(-(....)).....)))))).)))...... ( -23.80) >DroSim_CAF1 20544 85 + 1 GUGUCGGCCAACUA---AAAAAA--------------AAGCUAUCAAAGCGAAGAGGCUAGGAUGGGAAAAUGG-UUUUGGCAAAUAUUGGCCAACAGUUCAA .(((.((((((...---......--------------..(((.....)))......((((((((.(.....).)-))))))).....)))))).)))...... ( -22.40) >DroEre_CAF1 49691 84 + 1 GUGUCGGCCAACUA---AAA----------------AAACCCAUCAAAGAGAAGAGGCUAGGAUGGUAAAAUGGAUUUUGGCCAAUAUUGGCCAAUAGUUCAA ..............---...----------------....(((((..((........))..))))).....(((((((((((((....))))))).)))))). ( -21.00) >DroYak_CAF1 20560 99 + 1 GUGUCGGCCAACUA---AAAAAAAAACUAUAAAAAAAAACCAAUCAAAGAGAAGCGGCUAGGAUGGGAAAAUGG-UUUUGGCAAAUAUUGGCCAAUAGUUCAA .....((((((...---.....((((((((.........(((.((..((........))..)))))....))))-))))........)))))).......... ( -17.81) >DroAna_CAF1 32782 92 + 1 GUGUUGACAAACUGCUGAAA-AAACACAAAAAACAAAGACCAAUCGAGGGGA----GCUA-----GGGAAGUGG-CGUUGGCAAAUAUUGGCCAAUAGUUCAG (((((..((......))...-.)))))............((......))(((----((((-----......)))-)((((((........))))))..))).. ( -15.90) >consensus GUGUCGGCCAACUA___AAA_AA_____________AAACCUAUCAAAGCGAAGAGGCUAGGAUGGGAAAAUGG_UUUUGGCAAAUAUUGGCCAACAGUUCAA .(((.((((((............................((((((..((........))..))))))((((....))))........)))))).)))...... (-11.72 = -12.50 + 0.78)


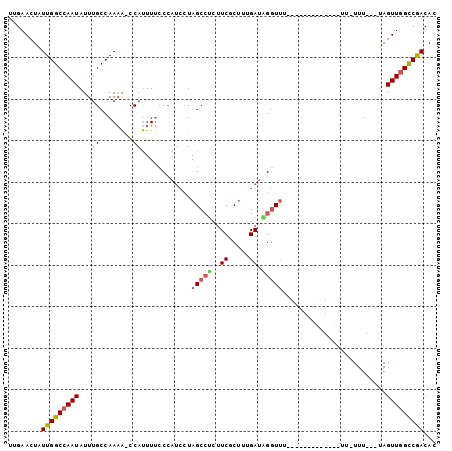
| Location | 22,902,860 – 22,902,956 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Mean single sequence MFE | -18.51 |
| Consensus MFE | -9.58 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22902860 96 - 27905053 UUGAACUGUUGGCCAAUAUUUGCCAAAA-CCAUUUUCCCAUCCUAGCCUCUUCGCUUUGAUAGGUUUUGUUUUU---UUUUUUUUU---UAGUUGGCCGACAC ......(((((((((((.......((((-.((....((.(((..(((......)))..))).))...)).))))---.........---..))))))))))). ( -22.17) >DroSec_CAF1 19863 82 - 1 UUGAACUGUUGGCCAAUAUUUGCCAAAA-CCAUUUUCCCAUCCUAGCCUCUUCGCUUUGAUAGGUU-----------------UUU---UAGUUGGCCGACAC ......(((((((((((.(((....)))-.......((.(((..(((......)))..))).))..-----------------...---..))))))))))). ( -21.50) >DroSim_CAF1 20544 85 - 1 UUGAACUGUUGGCCAAUAUUUGCCAAAA-CCAUUUUCCCAUCCUAGCCUCUUCGCUUUGAUAGCUU--------------UUUUUU---UAGUUGGCCGACAC ......(((((((((((.(((....)))-..........(((..(((......)))..))).....--------------......---..))))))))))). ( -19.10) >DroEre_CAF1 49691 84 - 1 UUGAACUAUUGGCCAAUAUUGGCCAAAAUCCAUUUUACCAUCCUAGCCUCUUCUCUUUGAUGGGUUU----------------UUU---UAGUUGGCCGACAC ........(((((((((..(((.......))).....(((((..((........))..)))))....----------------...---..)))))))))... ( -20.10) >DroYak_CAF1 20560 99 - 1 UUGAACUAUUGGCCAAUAUUUGCCAAAA-CCAUUUUCCCAUCCUAGCCGCUUCUCUUUGAUUGGUUUUUUUUUAUAGUUUUUUUUU---UAGUUGGCCGACAC ........(((((((((.......((((-(.((...........(((((..((.....)).))))).......)).))))).....---..)))))))))... ( -16.71) >DroAna_CAF1 32782 92 - 1 CUGAACUAUUGGCCAAUAUUUGCCAACG-CCACUUCCC-----UAGC----UCCCCUCGAUUGGUCUUUGUUUUUUGUGUUU-UUUCAGCAGUUUGUCAACAC (((((...(((((........)))))..-.(((.....-----.(((----...((......)).....)))....)))...-.))))).............. ( -11.50) >consensus UUGAACUAUUGGCCAAUAUUUGCCAAAA_CCAUUUUCCCAUCCUAGCCUCUUCGCUUUGAUAGGUUU_____________UU_UUU___UAGUUGGCCGACAC ........(((((((((...((........))............(((((..((.....)).))))).........................)))))))))... ( -9.58 = -10.00 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:24 2006