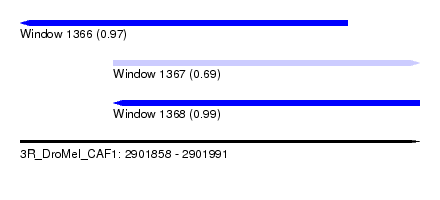
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,901,858 – 2,901,991 |
| Length | 133 |
| Max. P | 0.991979 |

| Location | 2,901,858 – 2,901,967 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -46.70 |
| Consensus MFE | -30.34 |
| Energy contribution | -31.34 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2901858 109 - 27905053 UCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA--GGCACAUUCCGCCCAACUCC---------CGCCUGGGGUGUAU (((((.....)))))..((((((.((((.((((.(((((....(((((......))))).)))))))))(((((.--(((.......)))...))))---------))))).)))))).. ( -50.60) >DroSec_CAF1 8611 109 - 1 UCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA--GGCACAUGCCGCCCAACUCC---------CGCCCGGGGUGUAU ..(((((((.((((((((((((....((((((((.........))))))))...)))..))))).))))..))))--)))((((.(((..(......---------.)..))).)))).. ( -47.80) >DroSim_CAF1 9627 109 - 1 UCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA--GGCACAUGCCGCCCAACUCC---------CGCCCGGGGUGUAU ..(((((((.((((((((((((....((((((((.........))))))))...)))..))))).))))..))))--)))((((.(((..(......---------.)..))).)))).. ( -47.80) >DroYak_CAF1 9548 105 - 1 UCGCCUCCUAAGCGAUGACCUCUGAGGCAAGUGCUGUAA---AGCAUUUGUAAUGAUGCGUCAUCCACUGG-GGA--GGCACUUUCAGCCCAACUCC---------CGCUCAGGGUGUAU ..(((((((.((.(((((((((....(((((((((....---)))))))))...)).).))))))..)).)-)))--)))............((.((---------(.....))).)).. ( -42.70) >DroAna_CAF1 8704 117 - 1 UCGCCAGAGAGGCGAUGACAUCUGCCAGAAGUGCUGUAG---AAUGUUUGUAAUAAUGCGUCAUCCGCUGGGGGGGUGGCAUUGUCAACCCGAUGCCCCAACCAGUCCCCGGGGGUGUAU (((((.....))))).(((((((((.((.....))))))---.))))).............(((((.((((((.((((((((((......)))))))...)))...)))))))))))... ( -44.60) >consensus UCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA__GGCACAUUCCGCCCAACUCC_________CGCCCGGGGUGUAU (((((.....)))))..((((((..(((.((((.(((((....(((((......))))).)))))))))((((....(((.......)))...))))..........)))..)))))).. (-30.34 = -31.34 + 1.00)

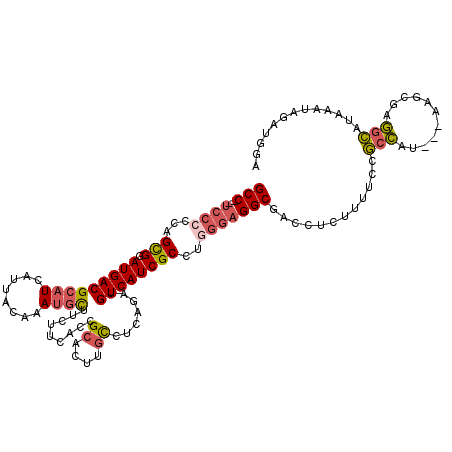
| Location | 2,901,889 – 2,901,991 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2901889 102 + 27905053 GCC--UCCCCCAGCGGAUGACGCAUCAUUACAAAUGCUUCUUCACCGCACUUGCCUCAGAUGUCAUCGCCUGGGAGGCGACCUCUUUUCCGCCAU---AAGCGAGGC------------- (((--((.(((((.(((((((((((........)))).........((....)).......)))))).)))))).((((..........))))..---....)))))------------- ( -35.60) >DroSec_CAF1 8642 115 + 1 GCC--UCCCCCAGCGGAUGACGCAUCAUUACAAAUGCUUCUUCACCGCACUUGCCUCAGAUGUCAUCGCCUGGGAGGCGACCUCUUUUCCGCCAU---AAGCGAGGCAUAAAUAGAUGGA (((--((.(((((.(((((((((((........)))).........((....)).......)))))).)))))).((((..........))))..---....)))))............. ( -37.30) >DroSim_CAF1 9658 115 + 1 GCC--UCCCCCAGCGGAUGACGCAUCAUUACAAAUGCUUCUUCACCGCACUUGCCUCAGAUGUCAUCGCCUGGGAGGCGACCUCUUUUCCGCCAU---AAGCGAGGCAUAAAUAGAUGGA (((--((.(((((.(((((((((((........)))).........((....)).......)))))).)))))).((((..........))))..---....)))))............. ( -37.30) >DroYak_CAF1 9579 110 + 1 GCC--UCC-CCAGUGGAUGACGCAUCAUUACAAAUGCU---UUACAGCACUUGCCUCAGAGGUCAUCGCUUAGGAGGCGACCUCUUUU-CGCCAU---AAUCAAGGCAUAAAUAGAUGGA (((--(((-..((((.(((((.(.((....(((.((((---....)))).))).....))))))))))))..))))))..(((((...-.(((..---......)))......))).)). ( -34.20) >DroAna_CAF1 8744 112 + 1 GCCACCCCCCCAGCGGAUGACGCAUUAUUACAAACAUU---CUACAGCACUUCUGGCAGAUGUCAUCGCCUCUCUGGCGACCUCUUUUCCACUUUCCCAAGUACAAUAUAAGUAG----- ............(((.....)))...............---((((........((((....))))(((((.....)))))...............................))))----- ( -18.20) >consensus GCC__UCCCCCAGCGGAUGACGCAUCAUUACAAAUGCUUCUUCACCGCACUUGCCUCAGAUGUCAUCGCCUGGGAGGCGACCUCUUUUCCGCCAU___AAGCGAGGCAUAAAUAGAUGGA (((..((((...(((.(((((((((........)))).........((....)).......))))))))..)))))))............(((...........)))............. (-19.16 = -19.84 + 0.68)

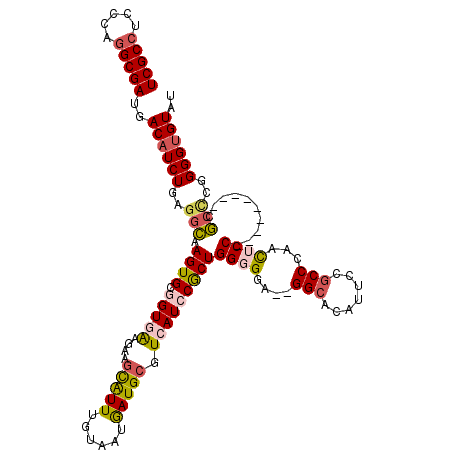
| Location | 2,901,889 – 2,901,991 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -42.82 |
| Consensus MFE | -27.58 |
| Energy contribution | -27.42 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2901889 102 - 27905053 -------------GCCUCGCUU---AUGGCGGAAAAGAGGUCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA--GGC -------------(((((.(((---..((((((.........(((.....)))((((.((((....((((((((.........))))))))...)))))))).))))))))).))--))) ( -45.70) >DroSec_CAF1 8642 115 - 1 UCCAUCUAUUUAUGCCUCGCUU---AUGGCGGAAAAGAGGUCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA--GGC .((.(((.(((.((((......---..)))).))))))))..(((((((.((((((((((((....((((((((.........))))))))...)))..))))).))))..))))--))) ( -47.10) >DroSim_CAF1 9658 115 - 1 UCCAUCUAUUUAUGCCUCGCUU---AUGGCGGAAAAGAGGUCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA--GGC .((.(((.(((.((((......---..)))).))))))))..(((((((.((((((((((((....((((((((.........))))))))...)))..))))).))))..))))--))) ( -47.10) >DroYak_CAF1 9579 110 - 1 UCCAUCUAUUUAUGCCUUGAUU---AUGGCG-AAAAGAGGUCGCCUCCUAAGCGAUGACCUCUGAGGCAAGUGCUGUAA---AGCAUUUGUAAUGAUGCGUCAUCCACUGG-GGA--GGC ...((((.(((.((((......---..))))-.))).)))).(((((((.((.(((((((((....(((((((((....---)))))))))...)).).))))))..)).)-)))--))) ( -42.60) >DroAna_CAF1 8744 112 - 1 -----CUACUUAUAUUGUACUUGGGAAAGUGGAAAAGAGGUCGCCAGAGAGGCGAUGACAUCUGCCAGAAGUGCUGUAG---AAUGUUUGUAAUAAUGCGUCAUCCGCUGGGGGGGUGGC -----.(((..(((((.(((..(.(....(((...(((.((((((.....))))))....))).)))....).).))).---)))))..))).......(((((((.(...).))))))) ( -31.60) >consensus UCCAUCUAUUUAUGCCUCGCUU___AUGGCGGAAAAGAGGUCGCCUCCCAGGCGAUGACAUCUGAGGCAAGUGCGGUGAAGAAGCAUUUGUAAUGAUGCGUCAUCCGCUGGGGGA__GGC ............(((((((((......))).....(((.((((((.....))))))....)))))))))((((.(((((....(((((......))))).)))))))))........... (-27.58 = -27.42 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:58 2006