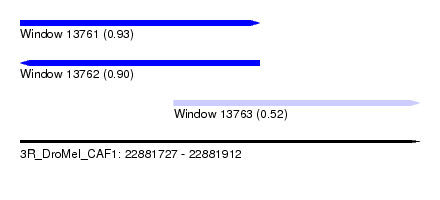
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,881,727 – 22,881,912 |
| Length | 185 |
| Max. P | 0.926515 |

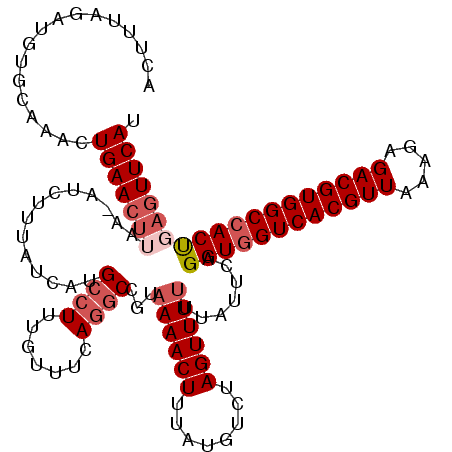
| Location | 22,881,727 – 22,881,838 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.87 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -24.56 |
| Energy contribution | -26.38 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22881727 111 + 27905053 AUUUUAGAUGUGCAAACUGAA---------UGUAUCAUGCCUUUGUUUCAGGCCGUUAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAU .......(((.((...(((((---------((......((((.......))))....(((((........)))))..)))))))(((((((((((.....)))))))))))...)).))) ( -31.70) >DroSec_CAF1 26725 120 + 1 ACUUUAGAUGUGCAAACUGAACUUAAGAUCUUUAUCAAGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAU .................(((((((..((..........((((.......))))...((((((........))))))....)).((((((((((((.....))))))))))))))))))). ( -32.30) >DroSim_CAF1 27905 119 + 1 ACUUUAGAUGUGAAAACUGAACUUAA-AUCUUUAUCAUGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUAGUCACGUUAAAGAGACGUGGCCACUGAGUUCAU ...........((((((((.((.(((-(..(((((...((((.......)))).)))))..)))).)).)))))))).((((((...((((((((.....))))))))..)))))).... ( -29.70) >DroYak_CAF1 31364 120 + 1 ACUCCAGAUGUGUAAGCUGAACUAAACAUCUCAAUCGUGGCUUUGUUUCAUGCCGUAAAACUUUAUGUCUAGUUUUUUAUUUAGGUGGUCACGUUAAAGAGACGUGGCCACCGAGUUCAU ((((....((.(((((..((((((.((((.....(..((((..........))))..)......)))).))))))))))).))((((((((((((.....)))))))))))))))).... ( -35.70) >consensus ACUUUAGAUGUGCAAACUGAACUUAA_AUCUUUAUCAUGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAU .................(((((((..............((((.......))))...((((((........)))))).......((((((((((((.....))))))))))))))))))). (-24.56 = -26.38 + 1.81)

| Location | 22,881,727 – 22,881,838 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.87 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -20.66 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22881727 111 - 27905053 AUGAACUCAGUGGCCACGUCUCUUUAACGUGACCACCUGAAUAAAAAACUAGACAUAAAGUUUAACGGCCUGAAACAAAGGCAUGAUACA---------UUCAGUUUGCACAUCUAAAAU .........((((.(((((.......))))).))))((((((.......(((((.....)))))...((((.......)))).......)---------)))))................ ( -24.50) >DroSec_CAF1 26725 120 - 1 AUGAACUCAGUGGCCACGUCUCUUUAACGUGACCACCUGAAUAAAAAACUAGACAUAAAGUUUUACGGCCUGAAACAAAGGCUUGAUAAAGAUCUUAAGUUCAGUUUGCACAUCUAAAGU ......(((((((.(((((.......))))).))).)))).....(((((..((.(((..((((..(((((.......))))).....))))..))).))..)))))............. ( -27.40) >DroSim_CAF1 27905 119 - 1 AUGAACUCAGUGGCCACGUCUCUUUAACGUGACUACCUGAAUAAAAAACUAGACAUAAAGUUUUACGGCCUGAAACAAAGGCAUGAUAAAGAU-UUAAGUUCAGUUUUCACAUCUAAAGU ......(((((((.(((((.......))))).))).))))....((((((..((.((((.(((((((((((.......)))).)).))))).)-))).))..))))))............ ( -26.80) >DroYak_CAF1 31364 120 - 1 AUGAACUCGGUGGCCACGUCUCUUUAACGUGACCACCUAAAUAAAAAACUAGACAUAAAGUUUUACGGCAUGAAACAAAGCCACGAUUGAGAUGUUUAGUUCAGCUUACACAUCUGGAGU ....(((((((((.(((((.......))))).))))).........((((((((((..((((....(((..........)))..))))...))))))))))...............)))) ( -32.30) >consensus AUGAACUCAGUGGCCACGUCUCUUUAACGUGACCACCUGAAUAAAAAACUAGACAUAAAGUUUUACGGCCUGAAACAAAGGCAUGAUAAAGAU_UUAAGUUCAGUUUGCACAUCUAAAGU .((((((((((((.(((((.......))))).))).))))....((((((........)))))).((((((.......)))).)).............)))))................. (-20.66 = -21.35 + 0.69)

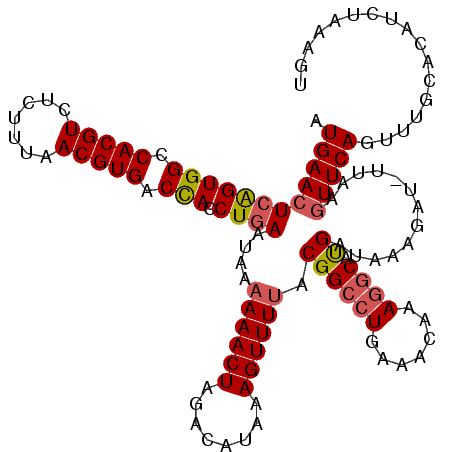
| Location | 22,881,798 – 22,881,912 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -22.46 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22881798 114 + 27905053 UCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUU------CAGGUCGAUUGUAUUUGCUUGCAGAAAAACAAUCCAAACU ((((.((((((((((.....))))))))))))))(((..((....(((((((.....(.(((............------..))).)......)))))))...))..))).......... ( -28.34) >DroSec_CAF1 26805 120 + 1 UCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCGGUUUCAGGUCGAUUGUAUCUGCUUGCAGAAAAACAAUCCAAACC ((((.((((((((((.....)))))))))))))).......(((........(((((((................))))).))...((((((.((((....))))...))))))..))). ( -32.19) >DroSim_CAF1 27984 120 + 1 UCAGGUAGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCGGUUUCAGGUCGAUUGUAUCUGCUUGCAGAAAAACAAUGCAAACU ((((...((((((((.....))))))))..))))((((((.(((........................(((((.(((.......)))))))).((((....))))..))).)))..))). ( -25.20) >DroYak_CAF1 31444 103 + 1 UUAGGUGGUCACGUUAAAGAGACGUGGCCACCGAGUUCAUUGUUAAAGUAAACUAAGCCACCAUAUUCUUAAAUCAGUUUCUGGUCGAUU---------U--------GCAAUCAAAAUU ...((((((((((((.....))))))))))))...............(((((.....(.((((.(..((......))..).)))).).))---------)--------)).......... ( -27.10) >consensus UCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCGGUUUCAGGUCGAUUGUAUCUGCUUGCAGAAAAACAAUCCAAACU ...((((((((((((.....))))))))))))((((.....(((..((....)).)))......))))..................((((((.((((....))))...))))))...... (-22.46 = -23.40 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:15 2006