
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,875,638 – 22,875,789 |
| Length | 151 |
| Max. P | 0.855754 |

| Location | 22,875,638 – 22,875,749 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -30.47 |
| Energy contribution | -30.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22875638 111 - 27905053 ACUUCUUCCUGCUCUGGUGGUGGCACGUGCACGUAGAUGUGCUUCUGCACCAAGGUGGUGCCACCACCACCACCGCCGCCGCCUCCGAAUCCGCCAAUGCUGCCACCUGUG ..........((...((((((((((((....)))....((((....))))...(((((((....)))))))...))))))))).........((....)).))........ ( -38.70) >DroWil_CAF1 27675 105 - 1 AUUUCCUCUUGCUCUGGUGGUGGGACAUGCACAUAGAUGUGCUUUUGCACUAGAGUGGUGCCACCACCA------CCGCCGCCUCCGAAACCACCUCCGAUGCUGCCUGUA ..............(((((((((.((..(((((....))))).....(((....))))).)))))))))------..((.((.((.((.......)).)).)).))..... ( -34.00) >DroMoj_CAF1 31113 105 - 1 ACUUCCUCCUGCUCUGGUGGUGGCACAUGCACAUAGAUGUGCUUCUGCACCAGAGUGGUGCCGCCA---CCGCCGCCGCCGCCUCCGAAUCCACCGCCUAUG---CCUGUA ..........((..((((((((((....(((((....)))))....(((((.....))))).))))---))))))..)).......................---...... ( -36.40) >consensus ACUUCCUCCUGCUCUGGUGGUGGCACAUGCACAUAGAUGUGCUUCUGCACCAGAGUGGUGCCACCACCACC_CCGCCGCCGCCUCCGAAUCCACCACCGAUGC__CCUGUA ...............(((((((((....(((((....)))))....(((((.....))))).............)))))))))............................ (-30.47 = -30.37 + -0.11)



| Location | 22,875,678 – 22,875,789 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.95 |
| Mean single sequence MFE | -42.94 |
| Consensus MFE | -33.84 |
| Energy contribution | -35.68 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22875678 111 + 27905053 GGUGGUGGUGGCACCACCUUGGUGCAGAAGCACAUCUACGUGCACGUGCCACCACCAGAGCAGGAAGAAGUGCGCCAGCGCCCUAACCUGCCCAUUGGCCAGUCCCAGAAG (((((((((((((((((....))).....((((......))))..))))))))))).(.(((((.....((((....)))).....))))).)....)))........... ( -49.80) >DroGri_CAF1 23297 111 + 1 GGCGGCGGUGGCACCACUUUGGUUCAAAAGCACAUUUAUGUUCAUGUGCCACCACCAGAGCAGGAGGAAGUGCGUCAGCGCCCCAACAUUCCAGUCGGCCAAUCCCAGAAG (((((.((((((((..((((......)))).(((....)))....)))))))).))......((((...((((....)))).......)))).....)))........... ( -33.70) >DroSec_CAF1 21409 111 + 1 GGUGGUGGUGGCACCACCUUGGUGCAGAAGCACAUCUACGUGCACGUGCCGCCACCAGAGCAGGAAGAAGUGCGCCAGCGCCCUAACCUGCCCAUUGGCCAGUCCCAGAAG (((((((((((((((((....))).....((((......))))..))))))))))).(.(((((.....((((....)))).....))))).)....)))........... ( -49.40) >DroWil_CAF1 27711 109 + 1 --UGGUGGUGGCACCACUCUAGUGCAAAAGCACAUCUAUGUGCAUGUCCCACCACCAGAGCAAGAGGAAAUCCGUCAGCGUCCCAACUUGCCAGUUGGUCAGUCUCAGAAG --(((((((((.(((((....))).....(((((....)))))..)).)))))))))((((..((((....)).))......((((((....))))))...).)))..... ( -37.40) >DroMoj_CAF1 31150 108 + 1 GG---UGGCGGCACCACUCUGGUGCAGAAGCACAUCUAUGUGCAUGUGCCACCACCAGAGCAGGAGGAAGUGCGUCAGCGACCUAACCUGCCCGUCGGCCAGGCCCAGAAG ((---((((((((((.....)))))....(((((....)))))...)))))))....(.((((((((...(((....))).)))..))))).)...(((...)))...... ( -44.40) >consensus GGUGGUGGUGGCACCACUUUGGUGCAGAAGCACAUCUAUGUGCAUGUGCCACCACCAGAGCAGGAGGAAGUGCGUCAGCGCCCUAACCUGCCCGUUGGCCAGUCCCAGAAG (((((((((((((((((....))).....((((......))))..)))))))))))...(((((..(..((((....))))..)..)))))......)))........... (-33.84 = -35.68 + 1.84)

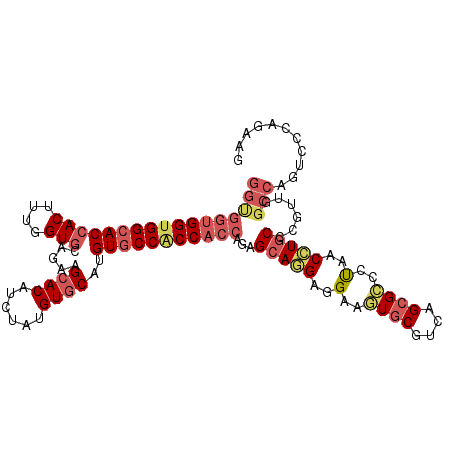

| Location | 22,875,678 – 22,875,789 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.95 |
| Mean single sequence MFE | -46.50 |
| Consensus MFE | -37.36 |
| Energy contribution | -38.64 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22875678 111 - 27905053 CUUCUGGGACUGGCCAAUGGGCAGGUUAGGGCGCUGGCGCACUUCUUCCUGCUCUGGUGGUGGCACGUGCACGUAGAUGUGCUUCUGCACCAAGGUGGUGCCACCACCACC ....(((......)))..(((((((..((((((....))).)))...)))))))((((((((((((..(((((....))))).....(((....))))))))))))))).. ( -53.60) >DroGri_CAF1 23297 111 - 1 CUUCUGGGAUUGGCCGACUGGAAUGUUGGGGCGCUGACGCACUUCCUCCUGCUCUGGUGGUGGCACAUGAACAUAAAUGUGCUUUUGAACCAAAGUGGUGCCACCGCCGCC .....((((..(((((((......))))).(((....)))....)))))).....(((((((((((....(((....)))(((((......))))).)))))))))))... ( -39.00) >DroSec_CAF1 21409 111 - 1 CUUCUGGGACUGGCCAAUGGGCAGGUUAGGGCGCUGGCGCACUUCUUCCUGCUCUGGUGGCGGCACGUGCACGUAGAUGUGCUUCUGCACCAAGGUGGUGCCACCACCACC ....(((......)))..(((((((..((((((....))).)))...)))))))((((((.(((((..(((((....))))).....(((....)))))))).)))))).. ( -49.70) >DroWil_CAF1 27711 109 - 1 CUUCUGAGACUGACCAACUGGCAAGUUGGGACGCUGACGGAUUUCCUCUUGCUCUGGUGGUGGGACAUGCACAUAGAUGUGCUUUUGCACUAGAGUGGUGCCACCACCA-- ......(((....((((((....))))))...((.((.((....))))..)))))((((((((.((..(((((....))))).....(((....))))).)))))))).-- ( -41.10) >DroMoj_CAF1 31150 108 - 1 CUUCUGGGCCUGGCCGACGGGCAGGUUAGGUCGCUGACGCACUUCCUCCUGCUCUGGUGGUGGCACAUGCACAUAGAUGUGCUUCUGCACCAGAGUGGUGCCGCCA---CC ......(((...)))...(((((((..((((.((....))))))...))))))).(((((((((((..(((((....)))))(((((...)))))..)))))))))---)) ( -49.10) >consensus CUUCUGGGACUGGCCAACGGGCAGGUUAGGGCGCUGACGCACUUCCUCCUGCUCUGGUGGUGGCACAUGCACAUAGAUGUGCUUCUGCACCAAAGUGGUGCCACCACCACC ....(((......)))..(((((((..((.(((....))).))....)))))))((((((((((((..(((((....))))).....(((....))))))))))))))).. (-37.36 = -38.64 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:08 2006