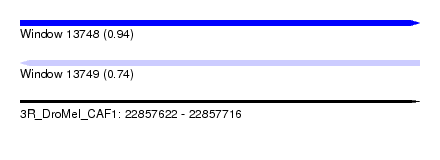
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,857,622 – 22,857,716 |
| Length | 94 |
| Max. P | 0.938925 |

| Location | 22,857,622 – 22,857,716 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.42 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22857622 94 + 27905053 UUUAAAGCUUUAACUUCUUGAGCAACUACUUGUGUUAUUUU--CCGA-----GACGG----AUUUUGGGGCUGUGGCAGCGUGCCAAAUGGCAUGUAAUGCUCAC ..................((((((.((((..((.(((...(--(((.-----..)))----)...))).)).))))..(((((((....)))))))..)))))). ( -28.90) >DroPse_CAF1 4039 80 + 1 GGUAUGGGUUUGGG------------------UUUGGUUUU--UGGG-----AUUGGGCCUAGGCUGGGUCUUUGGCAGCGUGCCAAAUGGCAUGUAAUGCUCAC ....(.((((((((------------------((..(((..--...)-----))..)))))))))).)......(((((((((((....)))))))..))))... ( -30.60) >DroSec_CAF1 3845 94 + 1 UUUAAAGCUUUAACUUCUUGAGCAACUACUUGUGUUAUUUU--UCGA-----GUCGG----AUUUUGGGGCUGUGGCAGCGUGCCAAAUGGCAUGUAAUGCUCAC ..................((((((........((((((.(.--.(((-----(....----..))))..)..))))))(((((((....)))))))..)))))). ( -26.80) >DroSim_CAF1 3864 94 + 1 UUUAAAGCUUUAACUUCUUGAGCAACUACUUGUGUUAUUUU--UCGA-----GGCGG----AUUUUGGGGCUGUGGCAGCGUGCCAAAUGGCAUGUAAUGCUCAC .....(((((((((((((((((.((..((....))..)).)--))))-----)).))----...))))))))(((((((((((((....)))))))..))).))) ( -27.30) >DroEre_CAF1 3895 101 + 1 CUUGAAGUUUUAACUUCUUAAGCAAUUUUCUGUGUUUUUUUUUUGGGCUGUUGGCGU----AUUUUGGGGCUGUGGCAGCGUGUCAAAUGGCAUGUAAUGCUCAC ((..((((.(((((..((((((.((.............)).))))))..)))))...----))))..))...(((((((((((((....)))))))..))).))) ( -21.82) >DroYak_CAF1 3744 96 + 1 UUUGAAGCUUUACCUUCUUAAGCAACUAUUUGUGUCUUUUUUUUUGG-----GACGG----AUUUUGGGGCUGUGGCAGCGUGCCAAAUGGCAUGUAAUGCUCAC ...((((......))))...(((..(....(.((((((.......))-----)))).----)....)..)))(((((((((((((....)))))))..))).))) ( -27.60) >consensus UUUAAAGCUUUAACUUCUUGAGCAACUACUUGUGUUAUUUU__UCGA_____GACGG____AUUUUGGGGCUGUGGCAGCGUGCCAAAUGGCAUGUAAUGCUCAC .....((((((((........((((....))))..........(((........))).......))))))))(((((((((((((....)))))))..))).))) (-17.48 = -17.42 + -0.06)

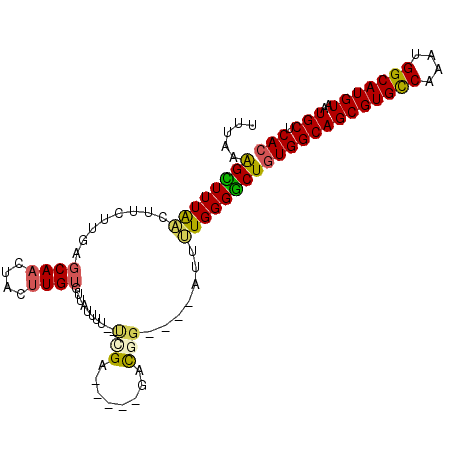
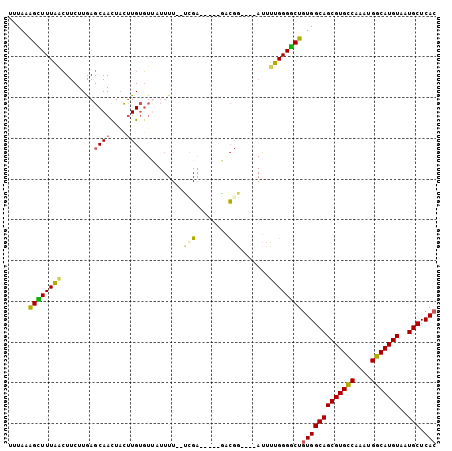
| Location | 22,857,622 – 22,857,716 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -18.75 |
| Consensus MFE | -10.33 |
| Energy contribution | -12.17 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22857622 94 - 27905053 GUGAGCAUUACAUGCCAUUUGGCACGCUGCCACAGCCCCAAAAU----CCGUC-----UCGG--AAAAUAACACAAGUAGUUGCUCAAGAAGUUAAAGCUUUAAA .((((((((((.((((....)))).((((...)))).......(----(((..-----.)))--)...........)))).)))))).(((((....)))))... ( -24.80) >DroPse_CAF1 4039 80 - 1 GUGAGCAUUACAUGCCAUUUGGCACGCUGCCAAAGACCCAGCCUAGGCCCAAU-----CCCA--AAAACCAAA------------------CCCAAACCCAUACC .((.((((...))))))((((((.....))))))......((....)).....-----....--.........------------------.............. ( -11.40) >DroSec_CAF1 3845 94 - 1 GUGAGCAUUACAUGCCAUUUGGCACGCUGCCACAGCCCCAAAAU----CCGAC-----UCGA--AAAAUAACACAAGUAGUUGCUCAAGAAGUUAAAGCUUUAAA .((((((((((.((((....)))).((((...))))........----.....-----....--............)))).)))))).(((((....)))))... ( -22.20) >DroSim_CAF1 3864 94 - 1 GUGAGCAUUACAUGCCAUUUGGCACGCUGCCACAGCCCCAAAAU----CCGCC-----UCGA--AAAAUAACACAAGUAGUUGCUCAAGAAGUUAAAGCUUUAAA .((((((((((.((((....)))).((((...))))........----.....-----....--............)))).)))))).(((((....)))))... ( -22.20) >DroEre_CAF1 3895 101 - 1 GUGAGCAUUACAUGCCAUUUGACACGCUGCCACAGCCCCAAAAU----ACGCCAACAGCCCAAAAAAAAAACACAGAAAAUUGCUUAAGAAGUUAAAACUUCAAG .((((((.....((...((((....((((...))))..))))..----..((.....))..............))......)))))).(((((....)))))... ( -13.60) >DroYak_CAF1 3744 96 - 1 GUGAGCAUUACAUGCCAUUUGGCACGCUGCCACAGCCCCAAAAU----CCGUC-----CCAAAAAAAAAGACACAAAUAGUUGCUUAAGAAGGUAAAGCUUCAAA .((((((..((.((((....)))).((((...))))........----..(((-----...........))).......)))))))).((((......))))... ( -18.30) >consensus GUGAGCAUUACAUGCCAUUUGGCACGCUGCCACAGCCCCAAAAU____CCGAC_____CCGA__AAAAUAACACAAGUAGUUGCUCAAGAAGUUAAAGCUUUAAA .((((((.....((((....)))).((((...)))).............................................)))))).((((......))))... (-10.33 = -12.17 + 1.83)

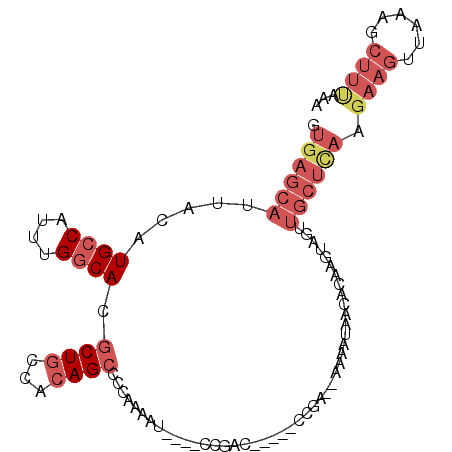
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:01 2006