
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
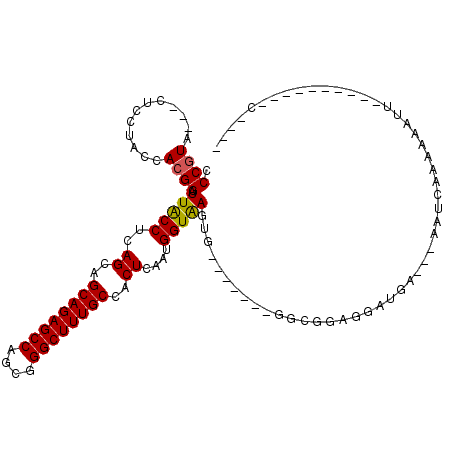
| Location | 22,820,732 – 22,820,837 |
| Length | 105 |
| Max. P | 0.834433 |

| Location | 22,820,732 – 22,820,837 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
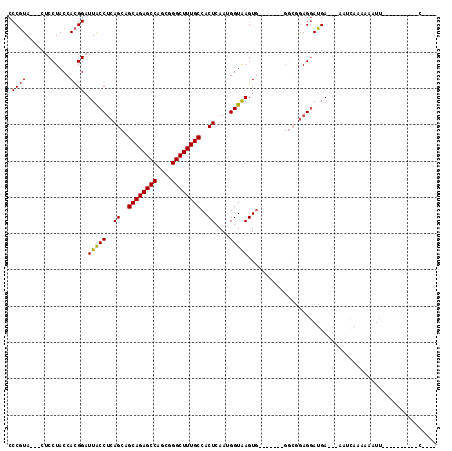
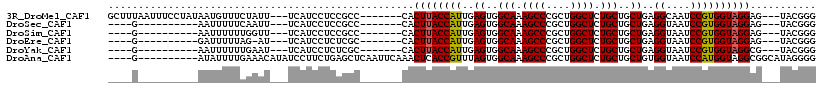
>3R_DroMel_CAF1 22820732 105 + 27905053 CCCGUA---CUCCUACCACGGAUUGCCUCAGCAGCAGAGCCAGCGGGCUUUGCCACUCAAUGGUAAGUG-------GGCGGAGGAUGA---AAUAGAAACAUUAUAGGAAAUUAAAGC .((...---((((..((((...(((((..((..((((((((....))))))))..))....))))))))-------)..))))((((.---........))))...)).......... ( -32.70) >DroSec_CAF1 78 91 + 1 CCCGUA---CUCCUACCACGGAUUACCUCAGCAGCAGAGCCAGCGGGCUUUGCCACUCAAUGGUAAGUG-------GGCGGAGGAUGA---AAUUGAAAAAUU----------C---- ..(((.---((((..((((...(((((..((..((((((((....))))))))..))....))))))))-------)..)))).))).---((((....))))----------.---- ( -32.30) >DroSim_CAF1 78 91 + 1 CCCGUA---CUCCUACCACGGAUUACCUCAGCAGCAGAGCCAGCGGGCUUUGCCACUCAAUGGUAAGUG-------GGCGGAGGAUGA---AACCAAAAAAUU----------C---- ..(((.---((((..((((...(((((..((..((((((((....))))))))..))....))))))))-------)..)))).))).---............----------.---- ( -32.20) >DroEre_CAF1 78 90 + 1 CCCGUA---CUCCUACCACGGAUUACCUCAGCAGCAGAGCCAGCGGGCUUUGCCACUCAAUGGUAAGUG-------GCGAGAGGAUGA---AU-CUAAAAAUC----------C---- .((((.---........))))....((((.((.((((((((....))))))))((((........))))-------))..))))....---..-.........----------.---- ( -27.70) >DroYak_CAF1 78 91 + 1 CCCGUA---CGCCUACCACGGAUUACCUCAGCAGCAGAGCCAGCGGGCUUUGCCACUCAAUGGUAAGUG-------GCGAGAGGAUGA---AUUCAAAAAAUU----------C---- .((((.---........))))....((((.((.((((((((....))))))))((((........))))-------))..))))....---............----------.---- ( -27.70) >DroAna_CAF1 9542 104 + 1 CCCCUAUGCCGCCUACCAUGGAUUACCACAGCAGCAGAGCCAGCGGGCUUUGCCACUAAACGGUGAGUUUGAAUUGAGCUCAGAAGGAUAUGUUUCAAAAUAU----------C---- ..(((..((((((......))........((..((((((((....))))))))..))...))))((((((.....))))))...)))................----------.---- ( -28.10) >consensus CCCGUA___CUCCUACCACGGAUUACCUCAGCAGCAGAGCCAGCGGGCUUUGCCACUCAAUGGUAAGUG_______GGCGGAGGAUGA___AAUCAAAAAAUU__________C____ .((((............)))).(((((..((..((((((((....))))))))..))....))))).................................................... (-20.54 = -20.43 + -0.11)

| Location | 22,820,732 – 22,820,837 |
|---|---|
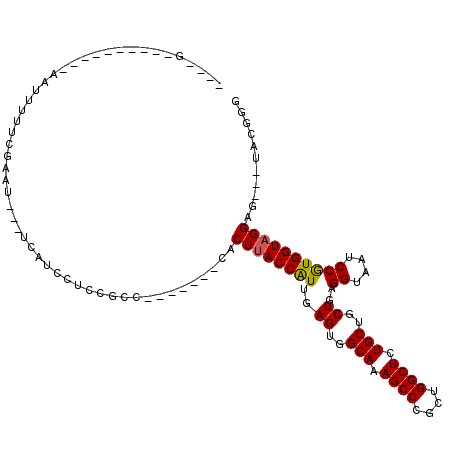
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -25.11 |
| Energy contribution | -25.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22820732 105 - 27905053 GCUUUAAUUUCCUAUAAUGUUUCUAUU---UCAUCCUCCGCC-------CACUUACCAUUGAGUGGCAAAGCCCGCUGGCUCUGCUGCUGAGGCAAUCCGUGGUAGGAG---UACGGG ...........................---.......(((..-------(.(((((((((.((..(((.((((....)))).)))..)).)((....)))))))))).)---..))). ( -30.30) >DroSec_CAF1 78 91 - 1 ----G----------AAUUUUUCAAUU---UCAUCCUCCGCC-------CACUUACCAUUGAGUGGCAAAGCCCGCUGGCUCUGCUGCUGAGGUAAUCCGUGGUAGGAG---UACGGG ----.----------............---.....((((..(-------((((((((..(.((..(((.((((....)))).)))..)).))))))...))))..))))---...... ( -31.50) >DroSim_CAF1 78 91 - 1 ----G----------AAUUUUUUGGUU---UCAUCCUCCGCC-------CACUUACCAUUGAGUGGCAAAGCCCGCUGGCUCUGCUGCUGAGGUAAUCCGUGGUAGGAG---UACGGG ----.----------........((..---....)).(((..-------(.(((((((((.((..(((.((((....)))).)))..)).)((....)))))))))).)---..))). ( -31.90) >DroEre_CAF1 78 90 - 1 ----G----------GAUUUUUAG-AU---UCAUCCUCUCGC-------CACUUACCAUUGAGUGGCAAAGCCCGCUGGCUCUGCUGCUGAGGUAAUCCGUGGUAGGAG---UACGGG ----(----------((((....)-))---))...((((..(-------((((((((..(.((..(((.((((....)))).)))..)).))))))...))))..))))---...... ( -33.90) >DroYak_CAF1 78 91 - 1 ----G----------AAUUUUUUGAAU---UCAUCCUCUCGC-------CACUUACCAUUGAGUGGCAAAGCCCGCUGGCUCUGCUGCUGAGGUAAUCCGUGGUAGGCG---UACGGG ----(----------(((((...))))---))..((...(((-------(...(((((((.((..(((.((((....)))).)))..)).)((....))))))))))))---...)). ( -32.80) >DroAna_CAF1 9542 104 - 1 ----G----------AUAUUUUGAAACAUAUCCUUCUGAGCUCAAUUCAAACUCACCGUUUAGUGGCAAAGCCCGCUGGCUCUGCUGCUGUGGUAAUCCAUGGUAGGCGGCAUAGGGG ----(----------((((........)))))((((((((...........))).((((((((..(((.((((....)))).)))..).((((....))))..)))))))...))))) ( -28.30) >consensus ____G__________AAUUUUUCGAAU___UCAUCCUCCGCC_______CACUUACCAUUGAGUGGCAAAGCCCGCUGGCUCUGCUGCUGAGGUAAUCCGUGGUAGGAG___UACGGG ...................................................((((((((..((..(((.((((....)))).)))..))..((....))))))))))........... (-25.11 = -25.00 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:55 2006