
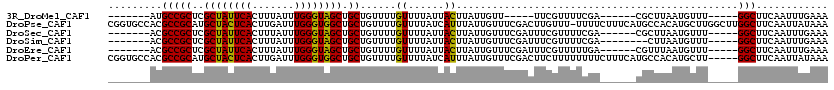
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,819,311 – 22,819,470 |
| Length | 159 |
| Max. P | 0.980605 |

| Location | 22,819,311 – 22,819,408 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -13.48 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22819311 97 + 27905053 -------AUGCCGCUCGCUAUUCACUUUAUUUGGGUAGCUGCUGUUUUGUUUUAUUACUUAUUGUU-----UUCGUUUUCGA------CGCUUAAUGUUU-----GGCUUCAAUUUGAAA -------..(((((..((((((((.......)))))))).)).................(((((..-----.(((....)))------....)))))...-----)))............ ( -16.40) >DroPse_CAF1 31472 119 + 1 CGGUGCCACGCCGCAUGCUACUCACUUGAUUUGGGUGGCUGCUGUUUUGUUUUAUCAUUUAUUGUUUCGACUUGUUU-UUUUCUUUCAUGCCACAUGCUUGGCUUGGCUUCAAUUAUAAA .((.((((.((((((.((((((((.......)))))))))))...................................-........((((...))))...))).)))).))......... ( -28.00) >DroSec_CAF1 28191 102 + 1 -------ACGCCGCUCGCUAUUCACUUUAUUUGGGUAGCUGCUGUUUUGUUUUAUUACUUAUUGUUUCGAUUUCGUUUUCGA------CGCUUAAUGUUU-----GGCUUCAAUUUGAAA -------..(((((..((((((((.......)))))))).)).....................((.((((........))))------.)).........-----)))............ ( -19.80) >DroSim_CAF1 28567 100 + 1 -------ACGCCGCUCGCUAUUCACUUUAUUUGGGUAGCUGCUGUUUUGUUUUAUUACUUAUUGUUUCGAUUUCGUUUUCGA--------CUUAAUGUUU-----GGCUUCAAUUUGAAA -------..(((((..((((((((.......)))))))).)).................(((((..((((........))))--------..)))))...-----)))............ ( -20.70) >DroEre_CAF1 28271 102 + 1 -------ACGCCGCUCGCUAUUCACUUUAUUUGGGUAGCUGCUGUUUUGUUUUAUUACUUAUUGUUUCGAUUUCGUUUUUGA------CGUUUAAUGUUU-----GGCUUCAAUUUGAAA -------..(((((..((((((((.......)))))))).)).................(((((..((((........))))------....)))))...-----)))............ ( -16.30) >DroPer_CAF1 31114 115 + 1 CGGUGCCACGCCGCAUGCUACUCACUUGAUUUGGGUGGCUGCUGUUUUGUUUUAUCAUUUAUUGUUUCGACUUCUUUUUUUUCUUUCAUGCCACAUGCUU-----GGCUUCAAUUAUAAA .((.((((.((.(((.((((((((.......)))))))))))......(((....((.....))....))).........................)).)-----))).))......... ( -23.70) >consensus _______ACGCCGCUCGCUAUUCACUUUAUUUGGGUAGCUGCUGUUUUGUUUUAUUACUUAUUGUUUCGAUUUCGUUUUCGA______CGCUUAAUGUUU_____GGCUUCAAUUUGAAA .........(((((..((((((((.......)))))))).))......((......))...............................................)))............ (-13.48 = -12.82 + -0.66)
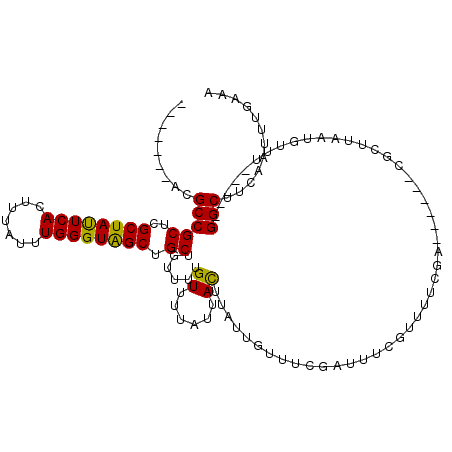

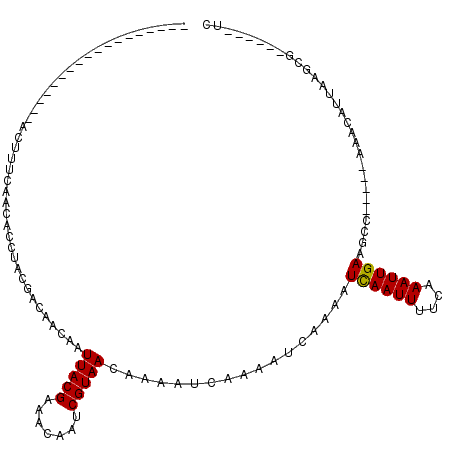
| Location | 22,819,379 – 22,819,470 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -11.62 |
| Consensus MFE | -5.52 |
| Energy contribution | -5.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22819379 91 - 27905053 ------------------ACUUUCAACACCUACGAUAACAAUUACGAAACAAUCGUAACAAAAUCAAAAUCAAAAUCAAUUUUCAAAUUGAAGCC-----AAACAUUAAGCG------UC ------------------.............(((.......((((((.....)))))).................((((((....))))))....-----..........))------). ( -8.00) >DroPse_CAF1 31551 120 - 1 AUCCACCAUUUACCAUCCACUUUCAACACCUACGACAACAAUUACGAAACAAACGUAACAAAAUCAAAAUCAAAAUCAAUUUUAUAAUUGAAGCCAAGCCAAGCAUGUGGCAUGAAAGAA ...................((((((................(((((.......))))).................((((((....)))))).((((.((...))...)))).)))))).. ( -17.20) >DroSec_CAF1 28264 91 - 1 ------------------ACUUUCAACACCUACGACAACAAUUACGAAACAAUCGUAACAAAAUCAAAAUCAAAAUUAAUUUUCAAAUUGAAGCC-----AAACAUUAAGCG------UC ------------------...............(((.....((((((.....)))))).................((((((....))))))....-----...........)------)) ( -7.00) >DroEre_CAF1 28344 91 - 1 ------------------ACUUUCAACACCUACGACAACAAUUACGAAACAAUCGUAACAAAAUCAAAAUCGAAAUCAAUUUUCAAAUUGAAGCC-----AAACAUUAAACG------UC ------------------...............(((.....((((((.....)))))).................((((((....))))))....-----...........)------)) ( -9.20) >DroPer_CAF1 31194 115 - 1 AUCCACCAUUUACCAUCCACUUUCAACACCUACGACAACAAUUACGAAACAAACGUAACAAAAUCAAAAUCAAAAUCAAUUUUAUAAUUGAAGCC-----AAGCAUGUGGCAUGAAAGAA ...................((((((................(((((.......))))).................((((((....)))))).(((-----(......)))).)))))).. ( -16.70) >consensus __________________ACUUUCAACACCUACGACAACAAUUACGAAACAAUCGUAACAAAAUCAAAAUCAAAAUCAAUUUUCAAAUUGAAGCC_____AAACAUUAAGCG______UC .........................................(((((.......))))).................((((((....))))))............................. ( -5.52 = -5.36 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:52 2006