
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,816,165 – 22,816,328 |
| Length | 163 |
| Max. P | 0.976823 |

| Location | 22,816,165 – 22,816,259 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22816165 94 + 27905053 UCAAAUCGGAACUGGUCAAGUGCAAGUGGCGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAGCUGACCGGCA-- ...........((((((...(((..((((.(((.....))).))))....)))...(((((((((.((......)).)))))))))))))))..-- ( -33.00) >DroSec_CAF1 25096 96 + 1 UCAAAUCGGAACUGGUCAAGUGCAAGUCGCGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAGCUGGCCGGCAAA .......((((((.(((..((((.....))))))).)))..)))....((((....(((((((((.((......)).))))))))).))))..... ( -28.30) >DroSim_CAF1 25463 96 + 1 UCAAAUCGGAACUGGUCAAGUGCAAGUCGCGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAGCUGGCCGGCAAA .......((((((.(((..((((.....))))))).)))..)))....((((....(((((((((.((......)).))))))))).))))..... ( -28.30) >DroEre_CAF1 25110 94 + 1 UCAAAUCGGAACUGGUCAAGUGCAAGUCGCGCGAUAAGUGCUCCAUAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAACUGGCCGGCA-- ...........(((((((.((....(..((((.....))))..)....((((((((((.((.......)).))))..)))))))))))))))..-- ( -23.80) >DroYak_CAF1 25293 94 + 1 UCAAAUCGGAACUGGUCAAGUGCAAGUUGGGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAUCUGGCCGGCA-- ...........(((((((.......((.(((((.....))))).)).(((((((((((.((.......)).))))..))))))).)))))))..-- ( -24.10) >consensus UCAAAUCGGAACUGGUCAAGUGCAAGUCGCGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAGCUGGCCGGCA__ ...........((((((..((((.....))))......((((.......))))...(((((((((.((......)).))))))))))))))).... (-24.66 = -25.10 + 0.44)


| Location | 22,816,187 – 22,816,289 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -18.99 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22816187 102 + 27905053 -----CAAGUGGCGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAGCU-GACCGGCA--------AACAGAGAUGACGGCCCACUA----AAGAGCCGA -----...((((.(((.....))).))))..((((....(((((((((.((......)).)))))))))-....(((.--------..((....))...))).....----....)))). ( -31.90) >DroPse_CAF1 28705 107 + 1 -----GUGGCCUGGCGAUAAGUGCUCCACAAUGGCAGAGUGCGGGCAAAUAUAUUUUUAAUUGCCAGUUAGGCUGGCA--------AACAGAAAUGACGGCCCACAAGCGCAAGAGUCCG -----(.(((((.(((....(..(((..(....)..)))..)((((.....(((((((..(((((((.....))))))--------)..)))))))...)))).....))).)).)))). ( -36.90) >DroSim_CAF1 25485 110 + 1 -----CAAGUCGCGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAGCU-GGCCGGCAAACAGGCAAACAGAGAUGACGGCCCACUA----AAGAGCCGA -----...(..((((.....))))..)....((((....(((((((((.((......)).)))))))))-(((((((......))...((....)).))))).....----....)))). ( -34.70) >DroEre_CAF1 25132 102 + 1 -----CAAGUCGCGCGAUAAGUGCUCCAUAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAACU-GGCCGGCA--------AACAGAGAUGACGGCCCACUA----AAGAGCCGA -----...(..((((.....))))..)....((((....((.((((((.((......)).)))))).))-(((((.((--------........)).))))).....----....)))). ( -27.00) >DroYak_CAF1 25315 102 + 1 -----CAAGUUGGGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAUCU-GGCCGGCA--------AACAGAGAUGACGGCCCACUA----AAGACCCGA -----....((((((....((((.......(((((((((((.((.......)).))))..)))))))..-(((((.((--------........)).))))))))).----..).))))) ( -26.80) >DroPer_CAF1 28350 112 + 1 AGGGAAGGGCCUGGCAAUAAGUGCUCCACAAUGGCAGAGUGCGGGCAAAUAUAUUUUUAAUUGCCAGUUAGGCUGGCA--------AACAGAAAUGACGGCCCACAAGCGCAAGAGUCCG ......((((((.((.....(..(((..(....)..)))..)((((.....(((((((..(((((((.....))))))--------)..)))))))...))))......)).)).)))). ( -39.20) >consensus _____CAAGUCGCGCGAUAAGUGCUCCACAAUGGCAAAGAGCUGGUAAAUACAUUUUUAAUUGCCAGCU_GGCCGGCA________AACAGAGAUGACGGCCCACUA____AAGAGCCGA .............(((.....)))........(((.....((((((((.((......)).))))))))..(((((......................))))).............))).. (-18.99 = -18.97 + -0.03)



| Location | 22,816,222 – 22,816,328 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
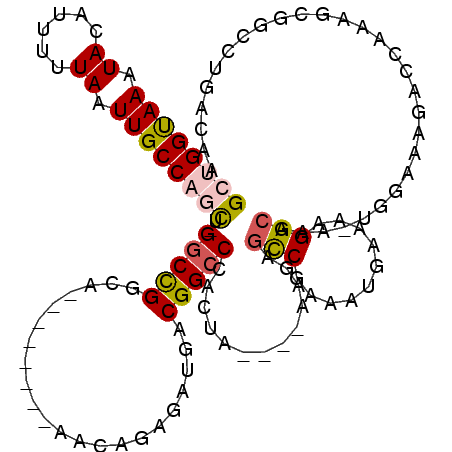
>3R_DroMel_CAF1 22816222 106 + 27905053 GCUGGUAAAUACAUUUUUAAUUGCCAGCU-GACCGGCA--------AACAGAGAUGACGGCCCACUA----AAGAGCCGAAAUGAAAAGGCAAA-UGGAAAGACCAACGCGGACGGACAA ((((((((.((......)).))))))))(-(.(((...--------...........((((.(....----..).))))..........((...-(((.....)))..))...))).)). ( -25.00) >DroPse_CAF1 28740 112 + 1 GCGGGCAAAUAUAUUUUUAAUUGCCAGUUAGGCUGGCA--------AACAGAAAUGACGGCCCACAAGCGCAAGAGUCCGAAUGAAAAGGCAAAACCAAAAGACUACAGCGGCCUGACAA ...(((((.((......)).))))).(((((((.(((.--------..((....))...)))......(((...((((....(....)((.....))....))))...)))))))))).. ( -32.10) >DroSim_CAF1 25520 101 + 1 GCUGGUAAAUACAUUUUUAAUUGCCAGCU-GGCCGGCAAACAGGCAAACAGAGAUGACGGCCCACUA----AAGAGCCGAAAUGAAAAGGCAAA-UGGAAAGACCAA------------- ((((((((.((......)).)))))))).-(((((((......))...((....)).))))).....----....(((..........)))...-(((.....))).------------- ( -27.40) >DroEre_CAF1 25167 106 + 1 GCUGGUAAAUACAUUUUUAAUUGCCAACU-GGCCGGCA--------AACAGAGAUGACGGCCCACUA----AAGAGCCGAAAUGAAAAGGCAAA-UGGAAAGACCUACGCGGCCUGACAA (((((((......((((((.(((((..((-(.......--------..)))......((((.(....----..).)))).........))))).-))))))....))).))))....... ( -25.10) >DroYak_CAF1 25350 106 + 1 GCUGGUAAAUACAUUUUUAAUUGCCAUCU-GGCCGGCA--------AACAGAGAUGACGGCCCACUA----AAGACCCGAAAUGAAAAGGCAAA-UGGAAAGACCAAAGCGGCCUGACAA ((((.(.......((((((.(((((....-(((((.((--------........)).))))).....----...........(....)))))).-))))))......).))))....... ( -22.52) >DroPer_CAF1 28390 112 + 1 GCGGGCAAAUAUAUUUUUAAUUGCCAGUUAGGCUGGCA--------AACAGAAAUGACGGCCCACAAGCGCAAGAGUCCGAAUGAAAAGGCAAAACGAAAAGACUACAGCGGCCUGACAA ...(((((.((......)).))))).(((((((.(((.--------..((....))...)))......(((...((((((..((......))...))....))))...)))))))))).. ( -30.90) >consensus GCUGGUAAAUACAUUUUUAAUUGCCAGCU_GGCCGGCA________AACAGAGAUGACGGCCCACUA____AAGAGCCGAAAUGAAAAGGCAAA_UGGAAAGACCAAAGCGGCCUGACAA ((((((((.((......)).))))))))..(((((......................))))).............(((..........)))............................. (-16.19 = -16.30 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:47 2006