
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,769,965 – 22,770,074 |
| Length | 109 |
| Max. P | 0.878349 |

| Location | 22,769,965 – 22,770,074 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
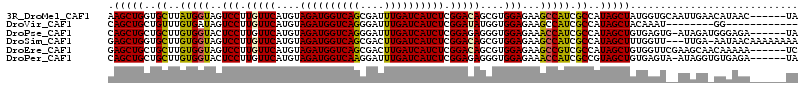
>3R_DroMel_CAF1 22769965 109 + 27905053 UA------GUUAUGUUCAAUUGCACCAUAGCUAUGGCGAUGGCUUCUCCACGCUGUCCGAGAUGAUCAAAUCGCUGACCAUCUACAUGAACAAGGACUACCAUAAGCACCAGCUU ((------(((.((((((...((......)).((((((.(((.....)))))))))...(((((.(((......))).)))))...))))))..)))))....((((....)))) ( -26.10) >DroVir_CAF1 10857 95 + 1 ------------CC--------AUUUGUAGCUAUGGCGAUGGCUUCUCCACCAUAUCCGAGAUGAUCAAAUCCCUGACCAUCUACAUGAACAAGGACUAUCACAAACAGCAGCUG ------------..--------....((.(((...(.(((((.((((....(((.....(((((.(((......))).)))))..)))....))))))))).)....))).)).. ( -18.50) >DroPse_CAF1 36164 108 + 1 UA------UCUCCCAUCUAU-CACUCACAGCUAUGGCGAUGGUUUCUCCACCCUCUCCGAGAUGAUCAAAUCCCUGACCAUCUACAUGAACAAGGAGUACCACAAGCAGCAGCUG ..------............-......(((((...((..((((..((((....((....(((((.(((......))).)))))....))....)))).))))...))...))))) ( -27.90) >DroSim_CAF1 5762 111 + 1 UUUUUUUUGUUAUU-UCAA---AACCAAAGCUAUGGCGAUGGCUUCUCCACGCUGUCCGAGAUGAUCAAGUCGCUGACCAUCUACAUGAACAAGGACUACCACAAGCACCAGCUC ....(((((.....-.)))---))....((((..((((.(((.....)))))))((((.(((((.(((......))).)))))..........)))).............)))). ( -22.70) >DroEre_CAF1 18145 109 + 1 GA------UUUUUGUUGCUUCGAACCACAGCUAUGGCGACGGCUUCUCCACGCUGUCCGAGAUGAUCAAGUCGCUGACCAUCUACAUGAACAAGGACUACCACAAGCAGCAGCUC ..------...(((((((((.(.........(((((.((((((........))))))).(((((.(((......))).))))).)))).....((....)).))))))))))... ( -27.60) >DroPer_CAF1 36180 108 + 1 UA------UCUCACACCUAU-UACUCACAGCUACGGCGAUGGUUUCUCCACCCUCUCCGAGAUGAUCAAAUCCUUGACCAUCUACAUGAACAAGGAGUACCACAAGCAGCAGCUG ..------............-......(((((.(.((..((((..((((....((....(((((.((((....)))).)))))....))....)))).))))...)).).))))) ( -28.70) >consensus UA______UUUAUC_UCAAU__AACCACAGCUAUGGCGAUGGCUUCUCCACCCUGUCCGAGAUGAUCAAAUCCCUGACCAUCUACAUGAACAAGGACUACCACAAGCAGCAGCUG ............................((((...((..(((.(..(((..........(((((.(((......))).)))))..........)))..))))...))...)))). (-17.02 = -17.22 + 0.19)



| Location | 22,769,965 – 22,770,074 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -25.69 |
| Energy contribution | -25.33 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22769965 109 - 27905053 AAGCUGGUGCUUAUGGUAGUCCUUGUUCAUGUAGAUGGUCAGCGAUUUGAUCAUCUCGGACAGCGUGGAGAAGCCAUCGCCAUAGCUAUGGUGCAAUUGAACAUAAC------UA ..(((.(((((.((((..((.(((.(((((((((((((((((....))))))))))......))))))).)))..))..))))))).)).).)).............------.. ( -31.30) >DroVir_CAF1 10857 95 - 1 CAGCUGCUGUUUGUGAUAGUCCUUGUUCAUGUAGAUGGUCAGGGAUUUGAUCAUCUCGGAUAUGGUGGAGAAGCCAUCGCCAUAGCUACAAAU--------GG------------ .(((..(((...(((((.(((((((..(((....)))..)))))))...)))))..))).(((((((..(....)..))))))))))......--------..------------ ( -28.70) >DroPse_CAF1 36164 108 - 1 CAGCUGCUGCUUGUGGUACUCCUUGUUCAUGUAGAUGGUCAGGGAUUUGAUCAUCUCGGAGAGGGUGGAGAAACCAUCGCCAUAGCUGUGAGUG-AUAGAUGGGAGA------UA ((((((..((..(((((.((((.(.(((.((.((((((((((....))))))))))))..))).).))))..))))).))..))))))......-............------.. ( -34.10) >DroSim_CAF1 5762 111 - 1 GAGCUGGUGCUUGUGGUAGUCCUUGUUCAUGUAGAUGGUCAGCGACUUGAUCAUCUCGGACAGCGUGGAGAAGCCAUCGCCAUAGCUUUGGUU---UUGA-AAUAACAAAAAAAA ((((..(.(((.((((..((.(((.(((((((((((((((((....))))))))))......))))))).)))..))..))))))).)..)))---)...-.............. ( -34.00) >DroEre_CAF1 18145 109 - 1 GAGCUGCUGCUUGUGGUAGUCCUUGUUCAUGUAGAUGGUCAGCGACUUGAUCAUCUCGGACAGCGUGGAGAAGCCGUCGCCAUAGCUGUGGUUCGAAGCAACAAAAA------UC ((((..(.(((.(((((..(.(((.(((((((((((((((((....))))))))))......))))))).)))..)..)))))))).)..)))).............------.. ( -38.10) >DroPer_CAF1 36180 108 - 1 CAGCUGCUGCUUGUGGUACUCCUUGUUCAUGUAGAUGGUCAAGGAUUUGAUCAUCUCGGAGAGGGUGGAGAAACCAUCGCCGUAGCUGUGAGUA-AUAGGUGUGAGA------UA (((((((.((..(((((.((((.(.(((.((.((((((((((....))))))))))))..))).).))))..))))).)).)))))))......-............------.. ( -38.10) >consensus CAGCUGCUGCUUGUGGUAGUCCUUGUUCAUGUAGAUGGUCAGCGAUUUGAUCAUCUCGGACAGCGUGGAGAAGCCAUCGCCAUAGCUGUGAUU__AUAGA_GAGAAA______UA .(((((..((..(((((..(((.(((((....((((((((((....)))))))))).)))))....)))...))))).))..)))))............................ (-25.69 = -25.33 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:26 2006