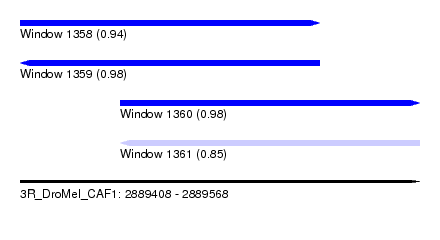
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,889,408 – 2,889,568 |
| Length | 160 |
| Max. P | 0.984373 |

| Location | 2,889,408 – 2,889,528 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -18.40 |
| Energy contribution | -21.07 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2889408 120 + 27905053 UAGCAAAUCUGCUCCAGUAGCUAAAACGGAUGUCAAAGAGAACAUCAAACAGGAGGUAAAGAAGGAGGUGAAGAAGGAGAUCGCUAAAACGCCAUCUCCGCCUCCUGCCCAACCACCAAA ((((....(((...)))..)))).....(((((........))))).....((.(((.....(((((((......((((((.((......)).))))))))))))).....))).))... ( -35.30) >DroSec_CAF1 1352 109 + 1 -----------CUCCAGUAGCUAAAACAAAUGUAAAAGAGAAUAUCAAACAGGAGGUAAAGAAGGAGGUGAAGAAGGAGAUCGCUAAAACGCCAUCUCCGCCUCCUGCCCAACCACCAAA -----------........................................((.(((.....(((((((......((((((.((......)).))))))))))))).....))).))... ( -27.90) >DroEre_CAF1 2925 108 + 1 AAGCAAAUCUGCACAAGUAGCUAAAGCGGCUGUAAAAGAGAAUAUCAAACAGGAGGUCA------------AGAAGGAGAUUGCUAAAACGCCACCUCCGCCUGCUCCCCAACCACCAAA .......(((.....(((.((....)).))).....)))............((.((...------------((..((((...((......))...))))..))...)))).......... ( -20.10) >consensus _AGCAAAUCUGCUCCAGUAGCUAAAACGGAUGUAAAAGAGAAUAUCAAACAGGAGGUAAAGAAGGAGGUGAAGAAGGAGAUCGCUAAAACGCCAUCUCCGCCUCCUGCCCAACCACCAAA ...................................................((.(((.....(((((((......((((((.((......)).))))))))))))).....))).))... (-18.40 = -21.07 + 2.67)

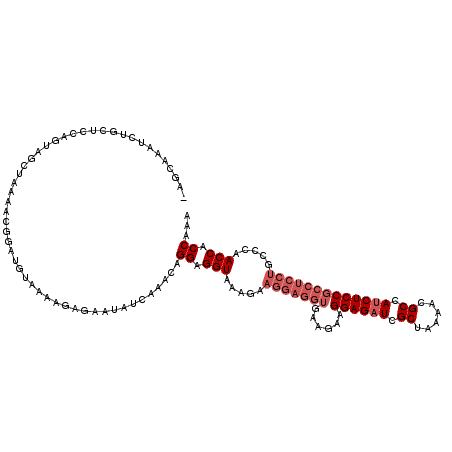
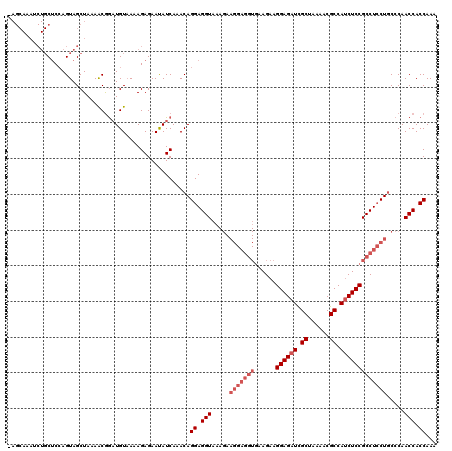
| Location | 2,889,408 – 2,889,528 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -26.26 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2889408 120 - 27905053 UUUGGUGGUUGGGCAGGAGGCGGAGAUGGCGUUUUAGCGAUCUCCUUCUUCACCUCCUUCUUUACCUCCUGUUUGAUGUUCUCUUUGACAUCCGUUUUAGCUACUGGAGCAGAUUUGCUA .(..((((((((((((((((.((((((.((......)).))))))...................))))))))..((((((......))))))....))))))))..)(((......))). ( -40.85) >DroSec_CAF1 1352 109 - 1 UUUGGUGGUUGGGCAGGAGGCGGAGAUGGCGUUUUAGCGAUCUCCUUCUUCACCUCCUUCUUUACCUCCUGUUUGAUAUUCUCUUUUACAUUUGUUUUAGCUACUGGAG----------- ((..(((((..(((((((((.((((((.((......)).))))))...................)))))))))..)......((...((....))...))))))..)).----------- ( -33.35) >DroEre_CAF1 2925 108 - 1 UUUGGUGGUUGGGGAGCAGGCGGAGGUGGCGUUUUAGCAAUCUCCUUCU------------UGACCUCCUGUUUGAUAUUCUCUUUUACAGCCGCUUUAGCUACUUGUGCAGAUUUGCUU ...(((((((((((((((((.((((((.((......)).))))))..))------------))..)))))....((.....)).....))))))))...((.......)).......... ( -31.30) >consensus UUUGGUGGUUGGGCAGGAGGCGGAGAUGGCGUUUUAGCGAUCUCCUUCUUCACCUCCUUCUUUACCUCCUGUUUGAUAUUCUCUUUUACAUCCGUUUUAGCUACUGGAGCAGAUUUGCU_ ((..(((((..(((((((((.((((((.((......)).))))))...................)))))))))..))................((....)))))..))............ (-26.26 = -26.82 + 0.56)

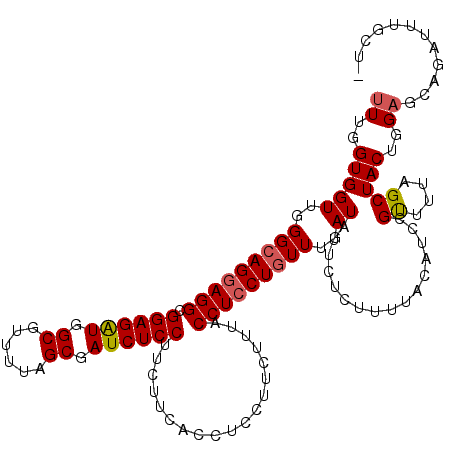
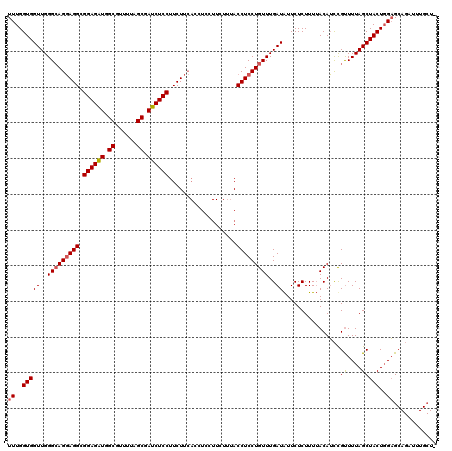
| Location | 2,889,448 – 2,889,568 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -18.40 |
| Energy contribution | -21.07 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
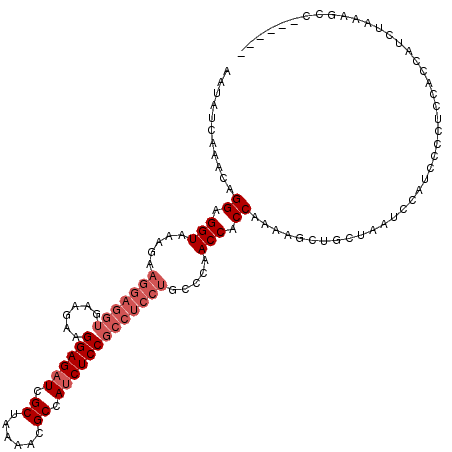
>3R_DroMel_CAF1 2889448 120 + 27905053 AACAUCAAACAGGAGGUAAAGAAGGAGGUGAAGAAGGAGAUCGCUAAAACGCCAUCUCCGCCUCCUGCCCAACCACCAAAAGCUGCUAAUCCAUCCCCUCCACCAUCUAAAGCCGCACCA ...........((.(((.....(((((((......((((((.((......)).))))))))))))).....))).))....((.(((.......................))).)).... ( -31.40) >DroSec_CAF1 1381 115 + 1 AAUAUCAAACAGGAGGUAAAGAAGGAGGUGAAGAAGGAGAUCGCUAAAACGCCAUCUCCGCCUCCUGCCCAACCACCAAAAGCUGCUAGUCCAUCCCCUCCACCAUCUAAAGCCC----- ...........((.(((.....(((((((......((((((.((......)).))))))))))))).....))).))....(((..(((.................))).)))..----- ( -30.13) >DroEre_CAF1 2965 96 + 1 AAUAUCAAACAGGAGGUCA------------AGAAGGAGAUUGCUAAAACGCCACCUCCGCCUGCUCCCCAACCACCAAAAGCUGCUAAUCCAUCCCC---ACCGCCAGCA--------- ...........((.((...------------((..((((...((......))...))))..))...))))...........(((((............---...).)))).--------- ( -17.26) >consensus AAUAUCAAACAGGAGGUAAAGAAGGAGGUGAAGAAGGAGAUCGCUAAAACGCCAUCUCCGCCUCCUGCCCAACCACCAAAAGCUGCUAAUCCAUCCCCUCCACCAUCUAAAGCC______ ...........((.(((.....(((((((......((((((.((......)).))))))))))))).....))).))........................................... (-18.40 = -21.07 + 2.67)

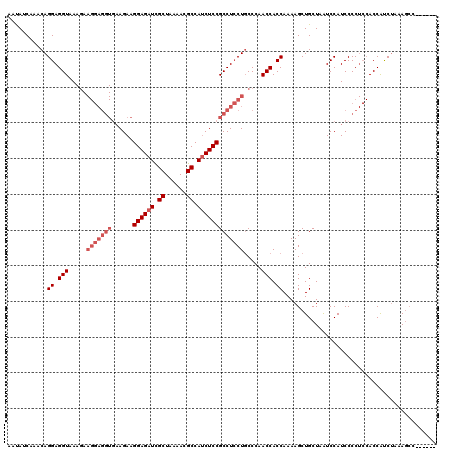
| Location | 2,889,448 – 2,889,568 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -28.45 |
| Energy contribution | -28.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
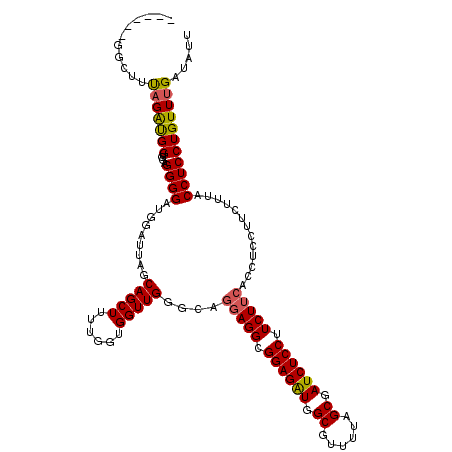
>3R_DroMel_CAF1 2889448 120 - 27905053 UGGUGCGGCUUUAGAUGGUGGAGGGGAUGGAUUAGCAGCUUUUGGUGGUUGGGCAGGAGGCGGAGAUGGCGUUUUAGCGAUCUCCUUCUUCACCUCCUUCUUUACCUCCUGUUUGAUGUU ....(((....(((..((((((((((..((.....(((((......)))))....(((((.((((((.((......)).)))))).))))).))..))))))))))..))).....))). ( -42.40) >DroSec_CAF1 1381 115 - 1 -----GGGCUUUAGAUGGUGGAGGGGAUGGACUAGCAGCUUUUGGUGGUUGGGCAGGAGGCGGAGAUGGCGUUUUAGCGAUCUCCUUCUUCACCUCCUUCUUUACCUCCUGUUUGAUAUU -----......(((..((((((((((..((.(((((.((.....)).)))))...(((((.((((((.((......)).)))))).))))).))..))))))))))..)))......... ( -42.90) >DroEre_CAF1 2965 96 - 1 ---------UGCUGGCGGU---GGGGAUGGAUUAGCAGCUUUUGGUGGUUGGGGAGCAGGCGGAGGUGGCGUUUUAGCAAUCUCCUUCU------------UGACCUCCUGUUUGAUAUU ---------.((.((.(((---.((((.......((.((((((.(....).))))))..))((((((.((......)).)))))).)))------------).))).)).))........ ( -30.50) >consensus ______GGCUUUAGAUGGUGGAGGGGAUGGAUUAGCAGCUUUUGGUGGUUGGGCAGGAGGCGGAGAUGGCGUUUUAGCGAUCUCCUUCUUCACCUCCUUCUUUACCUCCUGUUUGAUAUU ...........(((((((....((((.........(((((......)))))....(((((.((((((.((......)).)))))).))))).............)))))))))))..... (-28.45 = -28.57 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:50 2006