
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,716,152 – 22,716,294 |
| Length | 142 |
| Max. P | 0.969612 |

| Location | 22,716,152 – 22,716,254 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -45.65 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.74 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
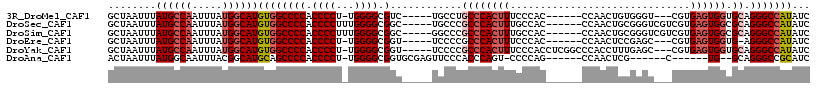
>3R_DroMel_CAF1 22716152 102 + 27905053 GCUAAUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCU-UGGGGCGUC-----UGCCUGCCCACUUUCCCAC------CCAACUGUGGGU---CGUGAGUGGUGCAGGGCCAUAUC ((........)).....((((((.....((((((.....-))))))...-----..(((((((((((.(((((------......))))).---...)))))).))))))))))).. ( -47.50) >DroSec_CAF1 6469 106 + 1 GCUAAUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUUUGGGGCGGC-----UGCCCGCCCACUUUGCCAC------CCAACUGCGGGUCGUCGUGAGUGGCGCAGGGCCAUAUC (((....(((((((.....)))))))..((((((......)))))))))-----.((((((((((((.((.((------((......)))).))...)))))).)).))))...... ( -48.70) >DroSim_CAF1 6393 106 + 1 GCUAAUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUUUGGGGCGGC-----GGCCCGCCCACUUUGCCAC------CCAACUGCGGGUCGUCGUGAGUGGCGCAGGGCCAUAUC (((....(((((((.....)))))))..((((((......)))))))))-----(((((((((((((.((.((------((......)))).))...)))))).)).)))))..... ( -53.70) >DroEre_CAF1 7778 101 + 1 GCUAAUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCU-UGGGGCGGU-----UCCCCGCCCACUUUCCCAC------CCAACUCCGAGC---CGUGAGUGGUG-AGGGCCAUAUC ........((((((.....))))))(((((((((((...-..((((((.-----...))))))..........------...((((((...---)).))))))))-.)))))))... ( -43.80) >DroYak_CAF1 6802 108 + 1 GCUAAUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCU-UGGGGCGGU-----UCCCCGCCCACUUUCCCACCUCGGCCCACCUUUGAGC---CGUGAGUGGUGCAGGGCCAUAUC ........((((((.....))))))(((((((.......-..((((((.-----...))))))......(((((.((((.((....)).))---)).).))))....)))))))... ( -47.30) >DroAna_CAF1 5387 95 + 1 ACUAAUUUAUGGCAAUUUACGGCAUGCAGCCCCACCCCU-UGGGGCGGUGCGAGUUCCCACCCAGU-CCCCAG------CCAACUCG------C------UG--GCAGGGCCGCAUC ...........((........))((((.((((((.....-))))))((((.(....).))))..((-((((((------(......)------)------))--)..)))).)))). ( -32.90) >consensus GCUAAUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCU_UGGGGCGGC_____UGCCCGCCCACUUUCCCAC______CCAACUGCGGGU___CGUGAGUGGUGCAGGGCCAUAUC ........((((((.....))))))((((((((.((((...)))).)............((((((((..............................)))))).)).)))))))... (-28.78 = -29.74 + 0.96)


| Location | 22,716,152 – 22,716,254 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -50.82 |
| Consensus MFE | -35.86 |
| Energy contribution | -36.83 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.20 |
| Mean z-score | -4.24 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22716152 102 - 27905053 GAUAUGGCCCUGCACCACUCACG---ACCCACAGUUGG------GUGGGAAAGUGGGCAGGCA-----GACGCCCCA-AGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAUUAGC ....(((((((((.(((((..(.---(((((....)))------)).)...))))))))....-----..(((((..-..))))))))))).((((((.....))))))........ ( -49.60) >DroSec_CAF1 6469 106 - 1 GAUAUGGCCCUGCGCCACUCACGACGACCCGCAGUUGG------GUGGCAAAGUGGGCGGGCA-----GCCGCCCCAAAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAUUAGC ....(((((((((.((.(((((..(.(((((....)))------)).)....))))).)))))-----)(((((((...)))))))))))).((((((.....))))))........ ( -52.80) >DroSim_CAF1 6393 106 - 1 GAUAUGGCCCUGCGCCACUCACGACGACCCGCAGUUGG------GUGGCAAAGUGGGCGGGCC-----GCCGCCCCAAAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAUUAGC ....((((((..((((((((((..((...))..).)))------)))))...(.((((((...-----.)))))))......)..)))))).((((((.....))))))........ ( -52.30) >DroEre_CAF1 7778 101 - 1 GAUAUGGCCCU-CACCACUCACG---GCUCGGAGUUGG------GUGGGAAAGUGGGCGGGGA-----ACCGCCCCA-AGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAUUAGC ....((((((.-((((.((((((---((.....)))..------)))))...(.((((((...-----.))))))).-...)))))))))).((((((.....))))))........ ( -48.60) >DroYak_CAF1 6802 108 - 1 GAUAUGGCCCUGCACCACUCACG---GCUCAAAGGUGGGCCGAGGUGGGAAAGUGGGCGGGGA-----ACCGCCCCA-AGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAUUAGC ....((((((..(.(((((..((---(((((....))))))).)))))....(.((((((...-----.))))))).-....)..)))))).((((((.....))))))........ ( -58.70) >DroAna_CAF1 5387 95 - 1 GAUGCGGCCCUGC--CA------G------CGAGUUGG------CUGGGG-ACUGGGUGGGAACUCGCACCGCCCCA-AGGGGUGGGGCUGCAUGCCGUAAAUUGCCAUAAAUUAGU .(((((((((..(--((------(------(......)------))((((-....((((.(....).)))).)))).-...))..)))))))))....................... ( -42.90) >consensus GAUAUGGCCCUGCACCACUCACG___ACCCGCAGUUGG______GUGGGAAAGUGGGCGGGCA_____ACCGCCCCA_AGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAUUAGC ....(((((((((.((..((((......(((....)))......))))......((((((.........))))))....)).))))))))).((((((.....))))))........ (-35.86 = -36.83 + 0.97)


| Location | 22,716,191 – 22,716,294 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -42.57 |
| Consensus MFE | -29.24 |
| Energy contribution | -29.92 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
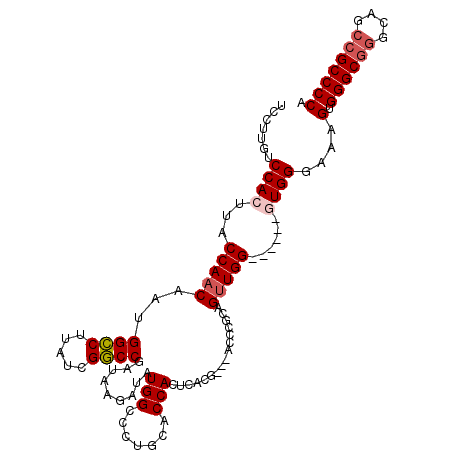
>3R_DroMel_CAF1 22716191 103 - 27905053 UCCUUGUCCACUUACCAACAAUGGCCUUAUCGGCCGAUAAGAUAUGGCCCUGCACCACUCACG---ACCCACAGUUGG------GUGGGAAAGUGGGCAGGCAGACGCCCCA ..((((((..............((((.....))))))))))....((((((((.(((((..(.---(((((....)))------)).)...)))))))))).....)))... ( -37.93) >DroSec_CAF1 6509 106 - 1 UGCUUGUCCACUUACCAACAAUGGCCUUAUCGGCCGAUAAGAUAUGGCCCUGCGCCACUCACGACGACCCGCAGUUGG------GUGGCAAAGUGGGCGGGCAGCCGCCCCA (((((((((((((.........((((.((((.........)))).))))....(((((((((..((...))..).)))------))))).)))))))))))))......... ( -44.80) >DroSim_CAF1 6433 106 - 1 UGCUUGUCCACUUACCAACAAAGGCCUUAUCGGCCGAUAAGAUAUGGCCCUGCGCCACUCACGACGACCCGCAGUUGG------GUGGCAAAGUGGGCGGGCCGCCGCCCCA .((((((((((((.........((((.((((.........)))).))))....(((((((((..((...))..).)))------))))).)))))))))))).......... ( -43.40) >DroEre_CAF1 7817 101 - 1 UCCGCGUCCAG-UACCAACAAUGGCCUUAUCGGCCGAUAAGAUAUGGCCCU-CACCACUCACG---GCUCGGAGUUGG------GUGGGAAAGUGGGCGGGGAACCGCCCCA (((.(..((((-..((.....(((((.....))))).........((((..-..........)---))).))..))))------..))))..(.((((((....))))))). ( -39.90) >DroYak_CAF1 6841 108 - 1 UCUGAGUCCAG-UACCAACAAUGGUCUUAUCGACCGAUAAGAUAUGGCCCUGCACCACUCACG---GCUCAAAGGUGGGCCGAGGUGGGAAAGUGGGCGGGGAACCGCCCCA ........(((-..(((......((((((((....)))))))).)))..)))..(((((..((---(((((....))))))).)))))....(.((((((....))))))). ( -46.80) >consensus UCCUUGUCCACUUACCAACAAUGGCCUUAUCGGCCGAUAAGAUAUGGCCCUGCACCACUCACG___ACCCGCAGUUGG______GUGGGAAAGUGGGCGGGCAGCCGCCCCA .......((((...(((((...((((.....)))).........(((.......)))................)))))......))))....(.((((((....))))))). (-29.24 = -29.92 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:00 2006