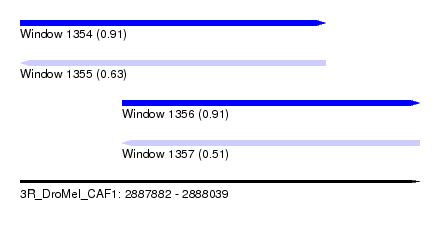
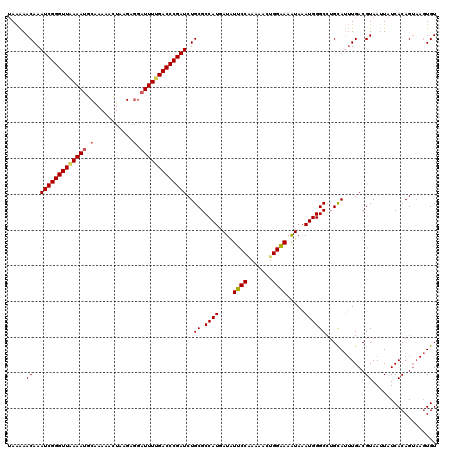
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,887,882 – 2,888,039 |
| Length | 157 |
| Max. P | 0.914487 |

| Location | 2,887,882 – 2,888,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
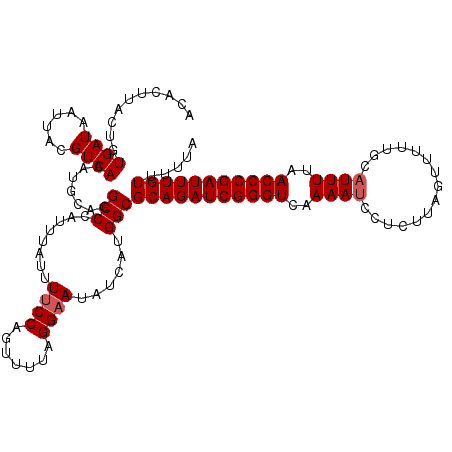
>3R_DroMel_CAF1 2887882 120 + 27905053 ACACUUACUGUGAUAAUUACGUCAAAUACAGGCCCAUUUACUUCCCAGUUUUCGGAACAUAAUGGCGCAGAUCGGGUCGAAAUCCUCUUAGUUUUUGCAUUUUAACCCGAUUUGUUCUUA .......(((((((......)))....))))(((.(((...((((........))))...))))))(((((((((((..((((.(...........).))))..)))))))))))..... ( -29.50) >DroEre_CAF1 1398 120 + 1 ACACUUACUGUGAUAUUUACGUCAAAUGCUGGCCCAUUUAUUUUCCAGUUUUUGGAAUAUCAUGGCGCAGAUCGGGUCAAAAUCCUCUUAGUUUUCGCCUUUUAACCCGAUUUGUUUUUA .......(((((((((((....((((.(((((............))))).))))))))))))))).(((((((((((.((((.................)))).)))))))))))..... ( -31.53) >DroYak_CAF1 21906 115 + 1 ACA----CUGUGAUAAUUACGUCA-UUGCAUGCCCAUUUAUUUUCCAGUUUUAGGAAUAUCAUGGCGCAGAUCGGGUCAAAAUCAUCCUAAUUUUUGCAUUUCAACCCGAUUUGUUUCUU ...----..(((((......))))-).....(((........((((.......))))......)))(((((((((((..((((((..........)).))))..)))))))))))..... ( -26.14) >consensus ACACUUACUGUGAUAAUUACGUCAAAUGCAGGCCCAUUUAUUUUCCAGUUUUAGGAAUAUCAUGGCGCAGAUCGGGUCAAAAUCCUCUUAGUUUUUGCAUUUUAACCCGAUUUGUUUUUA ..........((((......)))).......(((........((((.......))))......)))(((((((((((..((((...............))))..)))))))))))..... (-21.50 = -22.17 + 0.67)

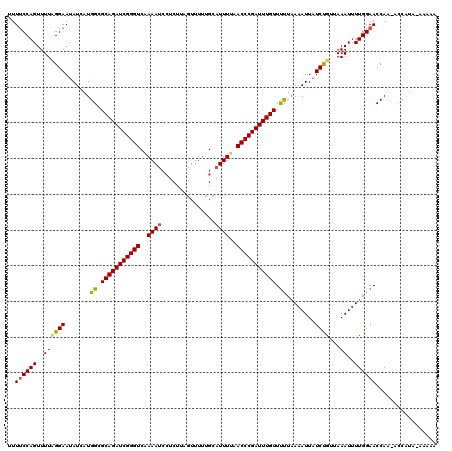
| Location | 2,887,882 – 2,888,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -22.33 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2887882 120 - 27905053 UAAGAACAAAUCGGGUUAAAAUGCAAAAACUAAGAGGAUUUCGACCCGAUCUGCGCCAUUAUGUUCCGAAAACUGGGAAGUAAAUGGGCCUGUAUUUGACGUAAUUAUCACAGUAAGUGU ((((.(((.(((((((..((((.(...........).))))..)))))))..((.(((((((.(((((.....))))).)).))))))).))).))))..........(((.....))). ( -26.70) >DroEre_CAF1 1398 120 - 1 UAAAAACAAAUCGGGUUAAAAGGCGAAAACUAAGAGGAUUUUGACCCGAUCUGCGCCAUGAUAUUCCAAAAACUGGAAAAUAAAUGGGCCAGCAUUUGACGUAAAUAUCACAGUAAGUGU .........((((((((((((..(...........)..))))))))))))(((.(((...((.(((((.....))))).)).....))))))((((((..((.......))..)))))). ( -29.10) >DroYak_CAF1 21906 115 - 1 AAGAAACAAAUCGGGUUGAAAUGCAAAAAUUAGGAUGAUUUUGACCCGAUCUGCGCCAUGAUAUUCCUAAAACUGGAAAAUAAAUGGGCAUGCAA-UGACGUAAUUAUCACAG----UGU ..((.....(((((((..((((.((..........))))))..))))))).(((.((((....((((.......)))).....))))))).....-...........))....----... ( -26.40) >consensus UAAAAACAAAUCGGGUUAAAAUGCAAAAACUAAGAGGAUUUUGACCCGAUCUGCGCCAUGAUAUUCCAAAAACUGGAAAAUAAAUGGGCCUGCAUUUGACGUAAUUAUCACAGUAAGUGU .....((..(((((((((((((.(...........).)))))))))))))..((.((((....((((.......)))).....))))))...........)).................. (-22.33 = -23.00 + 0.67)

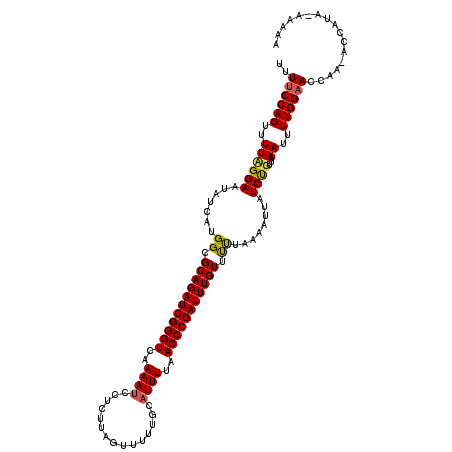
| Location | 2,887,922 – 2,888,039 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.19 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
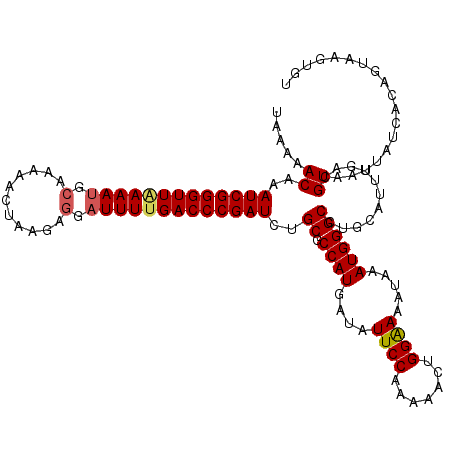
>3R_DroMel_CAF1 2887922 117 + 27905053 CUUCCCAGUUUUCGGAACAUAAUGGCGCAGAUCGGGUCGAAAUCCUCUUAGUUUUUGCAUUUUAACCCGAUUUGUUCUUAAAAUUAUCUGUUAAAUUUUGGAACCAA-AGCAUA--AAAA .((((.(((((.((((.......((.(((((((((((..((((.(...........).))))..))))))))))).))........))))..)))))..))))....-......--.... ( -27.16) >DroEre_CAF1 1438 119 + 1 UUUUCCAGUUUUUGGAAUAUCAUGGCGCAGAUCGGGUCAAAAUCCUCUUAGUUUUCGCCUUUUAACCCGAUUUGUUUUUAAAAUUAUCCGUUAAAUUUUGGAACCAA-ACCAUAAACAAA .......((((.(((...........(((((((((((.((((.................)))).)))))))))))((((((((((........))))))))))....-.))).))))... ( -25.63) >DroYak_CAF1 21941 120 + 1 UUUUCCAGUUUUAGGAAUAUCAUGGCGCAGAUCGGGUCAAAAUCAUCCUAAUUUUUGCAUUUCAACCCGAUUUGUUUCUUAAAUUAUCUGUAAAAUUUUGGAACCAAUACCUUACAAAAA ..(((((((((((.(.(((....((.(((((((((((..((((((..........)).))))..))))))))))).))......))).).))))))..)))))................. ( -26.10) >consensus UUUUCCAGUUUUAGGAAUAUCAUGGCGCAGAUCGGGUCAAAAUCCUCUUAGUUUUUGCAUUUUAACCCGAUUUGUUUUUAAAAUUAUCUGUUAAAUUUUGGAACCAA_ACCAUA_AAAAA ..((((((..((((((.......((.(((((((((((..((((...............))))..))))))))))).))........))))...))..))))))................. (-21.96 = -22.19 + 0.23)

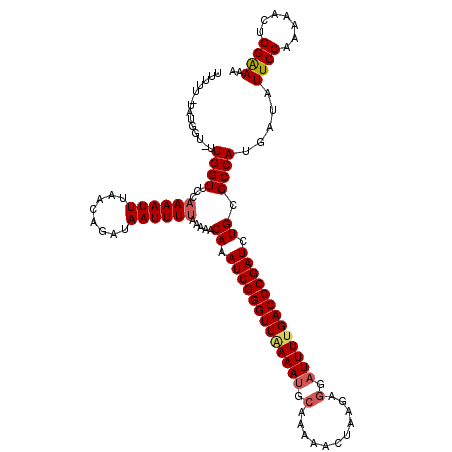
| Location | 2,887,922 – 2,888,039 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2887922 117 - 27905053 UUUU--UAUGCU-UUGGUUCCAAAAUUUAACAGAUAAUUUUAAGAACAAAUCGGGUUAAAAUGCAAAAACUAAGAGGAUUUCGACCCGAUCUGCGCCAUUAUGUUCCGAAAACUGGGAAG ....--...((.-...((((.((((((........))))))..))))..(((((((..((((.(...........).))))..)))))))..)).........((((........)))). ( -22.70) >DroEre_CAF1 1438 119 - 1 UUUGUUUAUGGU-UUGGUUCCAAAAUUUAACGGAUAAUUUUAAAAACAAAUCGGGUUAAAAGGCGAAAACUAAGAGGAUUUUGACCCGAUCUGCGCCAUGAUAUUCCAAAAACUGGAAAA .....(((((((-.....(((..........)))............((.((((((((((((..(...........)..)))))))))))).)).)))))))..(((((.....))))).. ( -28.20) >DroYak_CAF1 21941 120 - 1 UUUUUGUAAGGUAUUGGUUCCAAAAUUUUACAGAUAAUUUAAGAAACAAAUCGGGUUGAAAUGCAAAAAUUAGGAUGAUUUUGACCCGAUCUGCGCCAUGAUAUUCCUAAAACUGGAAAA ..((((((((((.(((....))).))))))))))............((.(((((((..((((.((..........))))))..))))))).))..........((((.......)))).. ( -29.40) >consensus UUUUU_UAUGGU_UUGGUUCCAAAAUUUAACAGAUAAUUUUAAAAACAAAUCGGGUUAAAAUGCAAAAACUAAGAGGAUUUUGACCCGAUCUGCGCCAUGAUAUUCCAAAAACUGGAAAA ..............((((...((((((........)))))).....((.(((((((((((((.(...........).))))))))))))).)).)))).....((((.......)))).. (-19.83 = -20.83 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:47 2006