
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,715,424 – 22,715,522 |
| Length | 98 |
| Max. P | 0.551082 |

| Location | 22,715,424 – 22,715,522 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -19.18 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
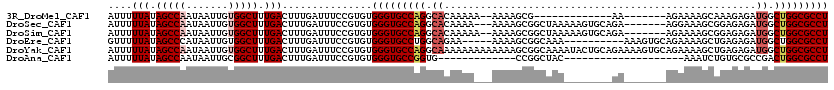
>3R_DroMel_CAF1 22715424 98 - 27905053 AUUUUUAUAGCCAAUAAUUGUGGCUUUGACUUUGAUUUCCGUGUGGGUGCCAGGCACAAAAA--AAAAGCG-------------AA-------AGAAAAGCAAAGAGAUGGCUGGCGCCU ....(((.(((((.......))))).)))...............(((((((((.((......--.....(.-------------..-------.).............)).))))))))) ( -25.95) >DroSec_CAF1 5733 110 - 1 AUUUUUAUAGCCAAUAAUUGUGGCUUUGACUUUGAUUUCCGUGUGGGUGCCAGGCACAAAA---AAAAGCGGCUAAAAAGUGCAGA-------AGGAAAGCGGAGAGAUGGCUGGCGCCU ....(((.(((((.......))))).)))(((((.(((((((.(((...))).))......---....(((.((....)))))...-------.))))).)))))....(((....))). ( -27.60) >DroSim_CAF1 5644 111 - 1 AUUUUUAUAGCCAAUAAUUGUGGCUUUGACUUUGAUUUCCGUGUGGGUGCCAGGCACAAAAA--AAAAGCGGCUAAAAAGUGCAGA-------AGAAAAGCGGAGAGAUGGCUGGCGCCU ....(((.(((((.......))))).)))...............(((((((((.(((.....--.......(((....)))((...-------......)).....).)).))))))))) ( -26.40) >DroEre_CAF1 6973 105 - 1 GUUUUUAUAGCCCAUAAUUGUGGCUUUGACUUUGAUUUCCGUGUGGGUGCCUGGCAGAA-----AAAAGCGGCAAA----------AAAGUGCAGAAAAGCUGAGAGAUGGCUGGCGCCU ....(((.((((((....)).)))).)))...............(((((((.(.((...-----...(((.(((..----------....)))......)))......)).).))))))) ( -27.90) >DroYak_CAF1 5997 120 - 1 AUUUUUAUAGCCAAUAAUUGUGGCUUUGACUUUGAUUUCCGUGUGGGUGCCAGGCAAAAAAAAAAAAAGCGGCAAAAUACUGCAGAAAAGUGCAGAAAAGCUGAGAGAUGGCUGGCGCCU ....(((.(((((.......))))).)))...............(((((((((.((.............((((......(((((......)))))....)))).....)).))))))))) ( -32.57) >DroAna_CAF1 4775 87 - 1 AUUUUUAUAGCCAAUAAUUGCGGCUUUGACUUUGAUUUCCGUGUGGGUGCCGGUG-------------CCGGCUAC--------------------AAAUCUGUGCGCCGACUGGCGCCU ....(((.((((.........)))).)))...............((((((((((.-------------.(((((((--------------------(....)))).)))))))))))))) ( -30.60) >consensus AUUUUUAUAGCCAAUAAUUGUGGCUUUGACUUUGAUUUCCGUGUGGGUGCCAGGCACAAAA___AAAAGCGGCUAA________GA_______AGAAAAGCUGAGAGAUGGCUGGCGCCU ....(((.(((((.......))))).)))...............(((((((((.((....................................................)).))))))))) (-19.18 = -19.90 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:58 2006