
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
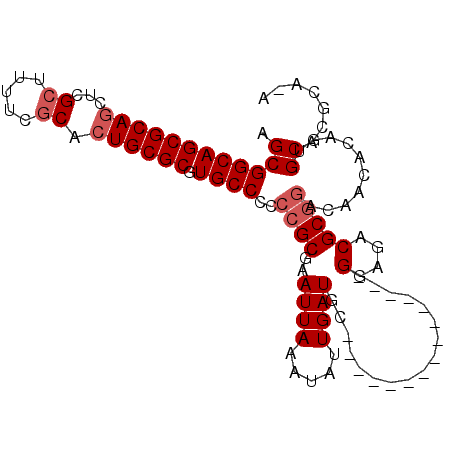
| Location | 22,692,839 – 22,692,936 |
| Length | 97 |
| Max. P | 0.934900 |

| Location | 22,692,839 – 22,692,936 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.58 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
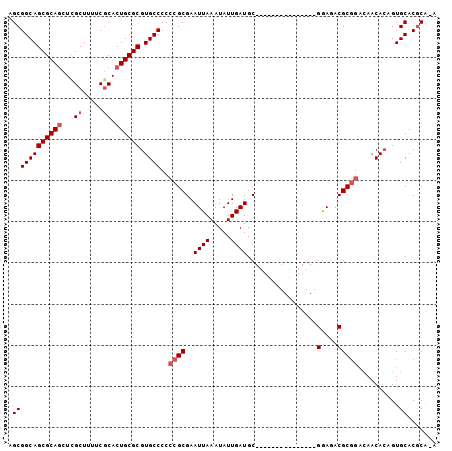
>3R_DroMel_CAF1 22692839 97 + 27905053 AGCGGCAGCGCAUCUCGCUUUUCCCACUGCGCGUGCCCCCCGCGAAUUAAAUAUUGAUGCGG-----GGAUUGC-GGAGACGCGGACAACAAAGUGCACGCA-A .((((.((((.....)))).......))))((((((.((((((((((.....)))..)))))-----)).((((-(....)))))..........)))))).-. ( -37.30) >DroSec_CAF1 140377 88 + 1 AGCGGCAGCGCAGCUCGCUUUUCGCACUGCGCGUGCCCCCCGCGAAUUAAAUAUUGAUGC---------------GAAGACGCGGACAACACAGUGCACGCG-A .((((((((((((...((.....)).))))))(((....(((((..((..((....))..---------------.))..)))))....)))..))).))).-. ( -30.50) >DroSim_CAF1 142563 88 + 1 AGCGGCAGCGCAGCUCGCUUUUCGCACUGCGCGUGCCCCCCGCGAAUUAAAUAUUGAUGC---------------GGAGACGCGGACAACACAGUGCACGCA-A .((((((((((((...((.....)).))))))(((....(((((.((((.....)))).)---------------(....)))))....)))..))).))).-. ( -31.70) >DroYak_CAF1 146295 99 + 1 AGCGGCAGCGCAGCUUGCUAUUCGCACUGCGCGUGCCCCCCGCGAAUUAAAUAUUGAUGCGUAUUCCGAAUUGCGGGAGACGC-----ACACAGUGCACUCGAA .((((((((((((..(((.....))))))))).))))...............((((.(((((.(((((.....))))).))))-----)..))))))....... ( -36.20) >consensus AGCGGCAGCGCAGCUCGCUUUUCGCACUGCGCGUGCCCCCCGCGAAUUAAAUAUUGAUGC_______________GGAGACGCGGACAACACAGUGCACGCA_A .((((((((((((...((.....)).)))))).))))..((((..((((.....)))).................(....)))))..........))....... (-22.58 = -23.58 + 1.00)

| Location | 22,692,839 – 22,692,936 |
|---|---|
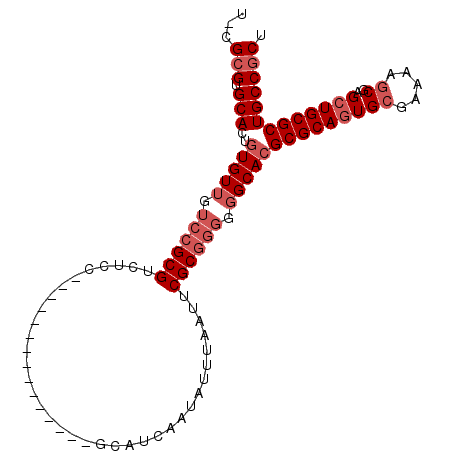
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -39.09 |
| Consensus MFE | -28.22 |
| Energy contribution | -30.22 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
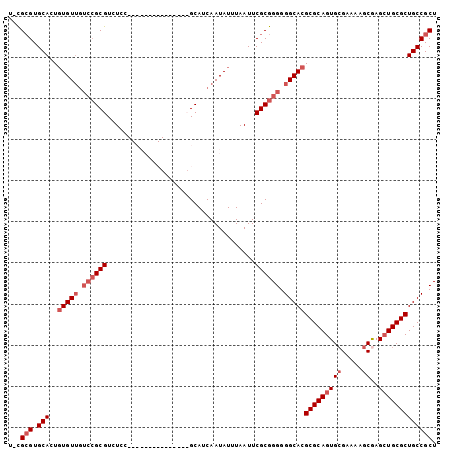
>3R_DroMel_CAF1 22692839 97 - 27905053 U-UGCGUGCACUUUGUUGUCCGCGUCUCC-GCAAUCC-----CCGCAUCAAUAUUUAAUUCGCGGGGGGCACGCGCAGUGGGAAAAGCGAGAUGCGCUGCCGCU .-.((((((............(((....)-))..(((-----((((...............)))))))))))))..(((((....((((.....)))).))))) ( -37.66) >DroSec_CAF1 140377 88 - 1 U-CGCGUGCACUGUGUUGUCCGCGUCUUC---------------GCAUCAAUAUUUAAUUCGCGGGGGGCACGCGCAGUGCGAAAAGCGAGCUGCGCUGCCGCU .-.(((.(((..(((((.((((((.....---------------................)))))).)))))(((((((.((.....)).))))))))))))). ( -38.80) >DroSim_CAF1 142563 88 - 1 U-UGCGUGCACUGUGUUGUCCGCGUCUCC---------------GCAUCAAUAUUUAAUUCGCGGGGGGCACGCGCAGUGCGAAAAGCGAGCUGCGCUGCCGCU .-.(((.(((..(((((.((((((.....---------------................)))))).)))))(((((((.((.....)).))))))))))))). ( -38.50) >DroYak_CAF1 146295 99 - 1 UUCGAGUGCACUGUGU-----GCGUCUCCCGCAAUUCGGAAUACGCAUCAAUAUUUAAUUCGCGGGGGGCACGCGCAGUGCGAAUAGCAAGCUGCGCUGCCGCU ......((((((((((-----(.(((((((((((((..(((((........))))))))).))))))))).)))))))))))...(((..((......)).))) ( -41.40) >consensus U_CGCGUGCACUGUGUUGUCCGCGUCUCC_______________GCAUCAAUAUUUAAUUCGCGGGGGGCACGCGCAGUGCGAAAAGCGAGCUGCGCUGCCGCU ...(((.(((..(((((.((((((....................................)))))).)))))(((((((((.....))..))))))))))))). (-28.22 = -30.22 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:46 2006