
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,644,664 – 22,644,762 |
| Length | 98 |
| Max. P | 0.634943 |

| Location | 22,644,664 – 22,644,762 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -20.04 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.56 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22644664 98 - 27905053 -----------------AACUC--CGCA-AGC--CGCAGUAAUGACAGUUCAAAAGGGCAACAAAGUACUAAAAAUAGUGGAAAUUACCGCAGGAUAUUCAAAUUUCCACAUUUGCGGAG -----------------..(((--((((-((.--...((((......((((....)))).......)))).......(((((((((...............))))))))).))))))))) ( -26.28) >DroVir_CAF1 123476 107 - 1 UGUGUGUUUAUAAUGUAAGCUC--CAUA--GU-----ACUACUGACAGUUCAAAAGGGCAACAAAGUACUAAAAAUAGUGGAAAUUACCGCAGGAUAUUCAAAUUUCCACAU-AAAG--- (((((((((((...))))))((--(.((--((-----(((..((...((((....))))..)).)))))))......((((......)))).))).............))))-)...--- ( -20.90) >DroGri_CAF1 169981 107 - 1 --GUAGUUU---AAGUAAGCUG--CAGA--GCAACACACUACUAACAGUUCAAAAGGGCAACAAAGUACUAAAAAUAGUGGAAAUUACCGCAGGAUAUUCAAAUUUCCACAU-AAAG--- --(((((((---....))))))--).((--((...............)))).....(....)...............(((((((((...............)))))))))..-....--- ( -18.42) >DroWil_CAF1 190243 91 - 1 -----------------AACUC--CGCAAAGC--CGCAGUAAUGACAGUUCAAAAGGGCAACAAAGUACUAAAAAUAGUGGAAAUUACCGCAGGAUAUUCAAAUUUCCACAC-------- -----------------.....--.((.....--.))((((......((((....)))).......)))).......(((((((((...............)))))))))..-------- ( -14.38) >DroMoj_CAF1 121073 95 - 1 ------------AUGUAAGCUC--CAUA--GU-----AGUACUGACAGUUCAAAAGGGCAACAAAGUACUAAAAAUAGUGGAAAUUACCGCAGGAUAUUCAAAUUUCCACAU-AAAG--- ------------..((((..((--(((.--.(-----((((((....((((....)))).....)))))))......)))))..))))....(((..........)))....-....--- ( -19.70) >DroPer_CAF1 105676 99 - 1 -----------------AUCCCCUCAGA-AGC--CACAGUAAUGACAGUUCAAAAGGGCAACAAAGUACUAAAAAUAGUGGAAAUUACCGCAGGAUAUUCAAAUUUCCACAU-UGCGGCG -----------------...........-.((--(.((((.......((((....))))..................(((((((((...............)))))))))))-)).))). ( -20.56) >consensus _________________AGCUC__CACA__GC__C_CAGUAAUGACAGUUCAAAAGGGCAACAAAGUACUAAAAAUAGUGGAAAUUACCGCAGGAUAUUCAAAUUUCCACAU_AAAG___ ...............................................((((....))))..................(((((((((...............))))))))).......... (-11.56 = -11.56 + -0.00)

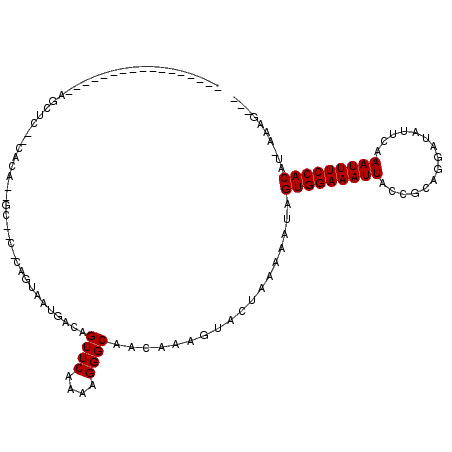
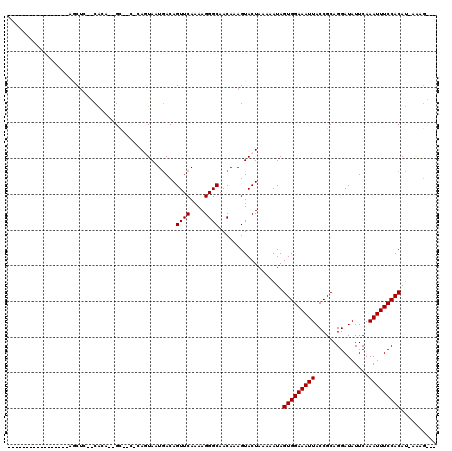
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:34 2006