
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,623,964 – 22,624,206 |
| Length | 242 |
| Max. P | 0.999548 |

| Location | 22,623,964 – 22,624,071 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.95 |
| Mean single sequence MFE | -17.87 |
| Consensus MFE | -12.66 |
| Energy contribution | -13.54 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22623964 107 + 27905053 ------UC----UUGCUCUACCACUUUCUCUUAUACUCUUAUACGAAGUACCUGUAUUUCAACUGUUUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGUUUAUAGUGCAACG ------..----((((.(((........................((((((....))))))...............(((((((.(.......))))))))........))).)))).. ( -19.20) >DroSec_CAF1 72141 113 + 1 GUUCUCUC----UUUCUCUACCACUUUCUCUUAUACUCUUAUACGAAGUACCUGUAUUUCAACUGUGUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGUUUAUAGUGCAACG (((.....----................................((((((....)))))).((((((..(.....(((((((.(.......))))))))...)..))))))..))). ( -17.40) >DroSim_CAF1 73952 113 + 1 CUUCUCUC----UUCCUCUACCACUUUCUCUUAUACUCUUAUACGAAGUACCUGUAUUUCAACUGUAUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUUUUUAUAGUGCAACG ........----................................((((((....)))))).((((((........(((((((.(.......))))))))......))))))...... ( -17.04) >DroEre_CAF1 80324 113 + 1 ---CUCUCUUCCUCUCUCUGAAACAUUCUCUUAUACUCGUGUACAAAGUACCUGUAUUUCAAAUGUAUUGCUCUCUCGCAUGACU-UUAUCGCAUGCGAUUUGCUUAUAGUGCAACG ---..................................(((((((.(((((....)))))....((((..((....(((((((.(.-.....))))))))...)).)))))))).))) ( -18.10) >DroYak_CAF1 75417 110 + 1 ---CUCUC----UCUCUCAGCCACUUUCUCUUAUACGCGUAUACGAAGUACCUGUAUUACAACUGUAUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGCUUAUAGCGCAACG ---.....----........................((((.....(((((.......(((....)))........(((((((.(.......))))))))..)))))...)))).... ( -17.60) >consensus ___CUCUC____UUUCUCUACCACUUUCUCUUAUACUCUUAUACGAAGUACCUGUAUUUCAACUGUAUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGUUUAUAGUGCAACG ............................................((((((....)))))).((((((........(((((((.(.......))))))))......))))))...... (-12.66 = -13.54 + 0.88)

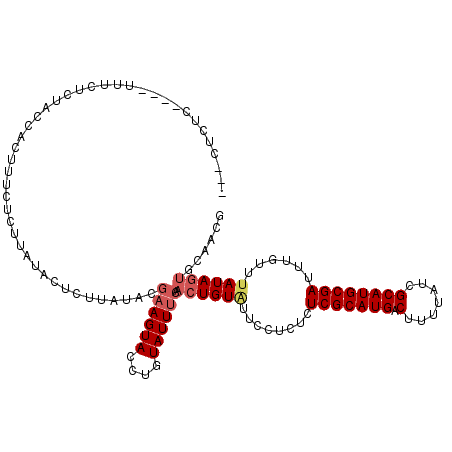
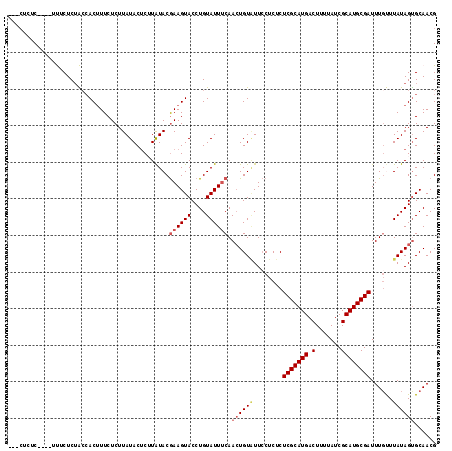
| Location | 22,623,991 – 22,624,099 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 94.54 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22623991 108 + 27905053 CUUAUACGAAGUACCUGUAUUUCAACUGUUUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGUUUAUAGUGCAACGACCACAGACACGAAGAUAAUUCGUGUGU .......((((((....))))))..((((..((.....(((((((.(.......)))))))).((((.......)))).))..))))(((((((.....))))))).. ( -26.60) >DroSec_CAF1 72174 108 + 1 CUUAUACGAAGUACCUGUAUUUCAACUGUGUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGUUUAUAGUGCAACGACCACAGACACGAAGAUAAUUCGUGUGU .......((((((....))))))..(((((.((.....(((((((.(.......)))))))).((((.......)))).)).)))))(((((((.....))))))).. ( -30.10) >DroSim_CAF1 73985 108 + 1 CUUAUACGAAGUACCUGUAUUUCAACUGUAUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUUUUUAUAGUGCAACGACCACAGACACGAAGAUAAUUCGUGUGU .......((((((....)))))).((((((........(((((((.(.......))))))))......)))))).............(((((((.....))))))).. ( -25.34) >DroEre_CAF1 80358 107 + 1 CGUGUACAAAGUACCUGUAUUUCAAAUGUAUUGCUCUCUCGCAUGACU-UUAUCGCAUGCGAUUUGCUUAUAGUGCAACGACCACAGACACGAAGAUAAUUCGUGUAU (((((((.(((((....)))))....((((..((....(((((((.(.-.....))))))))...)).)))))))).))).......(((((((.....))))))).. ( -27.40) >DroYak_CAF1 75447 108 + 1 CGUAUACGAAGUACCUGUAUUACAACUGUAUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGCUUAUAGCGCAACGACCACAGACACGAAGAUUAUUCGUGUGU .(((((((.......)))).)))..((((..((.....(((((((.(.......)))))))).((((.......)))).))..))))(((((((.....))))))).. ( -25.90) >consensus CUUAUACGAAGUACCUGUAUUUCAACUGUAUUCCUCUCUCGCAUGACUUUUAUCGCAUGCGAUUUGUUUAUAGUGCAACGACCACAGACACGAAGAUAAUUCGUGUGU .......((((((....))))))..((((.........(((((((.(.......)))))))).((((.......)))).....))))(((((((.....))))))).. (-22.74 = -23.30 + 0.56)

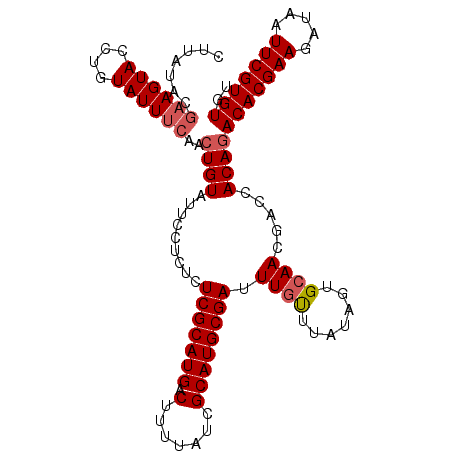
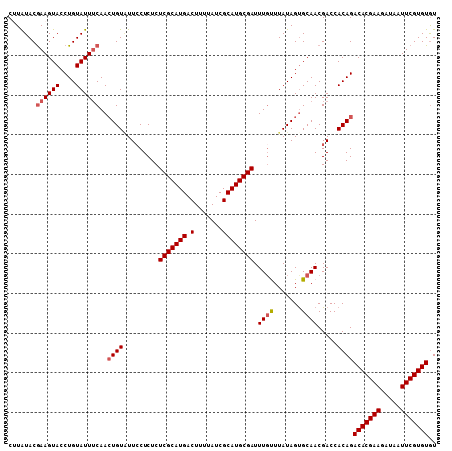
| Location | 22,623,991 – 22,624,099 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.54 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -22.60 |
| Energy contribution | -22.44 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22623991 108 - 27905053 ACACACGAAUUAUCUUCGUGUCUGUGGUCGUUGCACUAUAAACAAAUCGCAUGCGAUAAAAGUCAUGCGAGAGAGGAAAACAGUUGAAAUACAGGUACUUCGUAUAAG ....(((((.((((((((...((((..(((((........)))...((((((((.......).))))))).....))..)))).))).....))))).)))))..... ( -26.10) >DroSec_CAF1 72174 108 - 1 ACACACGAAUUAUCUUCGUGUCUGUGGUCGUUGCACUAUAAACAAAUCGCAUGCGAUAAAAGUCAUGCGAGAGAGGAACACAGUUGAAAUACAGGUACUUCGUAUAAG ....(((((.((((((((...(((((.(((((........)))...((((((((.......).))))))).....)).))))).))).....))))).)))))..... ( -27.60) >DroSim_CAF1 73985 108 - 1 ACACACGAAUUAUCUUCGUGUCUGUGGUCGUUGCACUAUAAAAAAAUCGCAUGCGAUAAAAGUCAUGCGAGAGAGGAAUACAGUUGAAAUACAGGUACUUCGUAUAAG ....(((((.((((((((...(((((.((.((..............((((((((.......).)))))))..)).)).))))).))).....))))).)))))..... ( -24.39) >DroEre_CAF1 80358 107 - 1 AUACACGAAUUAUCUUCGUGUCUGUGGUCGUUGCACUAUAAGCAAAUCGCAUGCGAUAA-AGUCAUGCGAGAGAGCAAUACAUUUGAAAUACAGGUACUUUGUACACG ..(((((((.....)))))))..(((((.(((((.((.........((((((((.....-.).))))))).)).)))))))........(((((.....)))))))). ( -26.60) >DroYak_CAF1 75447 108 - 1 ACACACGAAUAAUCUUCGUGUCUGUGGUCGUUGCGCUAUAAGCAAAUCGCAUGCGAUAAAAGUCAUGCGAGAGAGGAAUACAGUUGUAAUACAGGUACUUCGUAUACG ..(((((((.....)))))))(((((.((.((((.......)))).((((((((.......).))))))).....)).)))))..(((.(((.((....)))))))). ( -27.50) >consensus ACACACGAAUUAUCUUCGUGUCUGUGGUCGUUGCACUAUAAACAAAUCGCAUGCGAUAAAAGUCAUGCGAGAGAGGAAUACAGUUGAAAUACAGGUACUUCGUAUAAG ....(((((.(((((...(((.((((((......)))))).)))..((((((((.......).)))))))......................))))).)))))..... (-22.60 = -22.44 + -0.16)

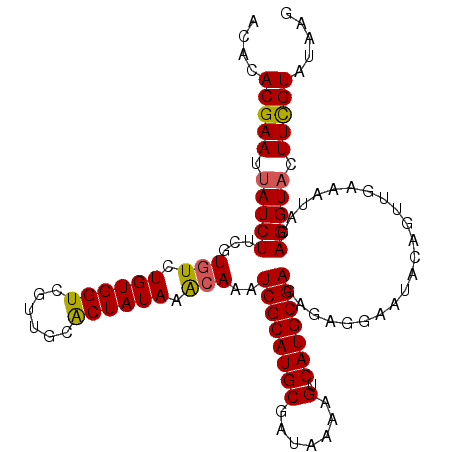
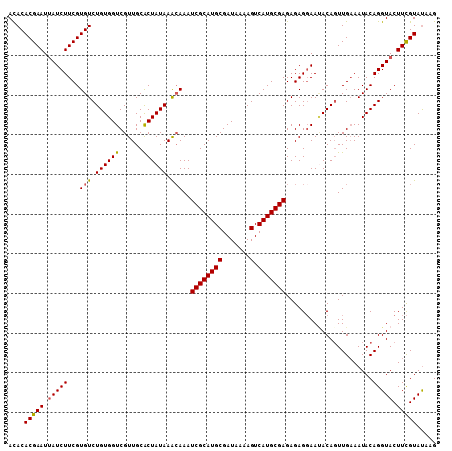
| Location | 22,624,071 – 22,624,170 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.61 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22624071 99 + 27905053 ACCACAGACACGAAGAUAAUUCGUGUGUCUCUCUCUUUUUCUCCCACGCAUCUGUACCUUCGAACGGUAUUUGUGGGGUAGAAGAAAUUGCUCGACUUA .......(((((((.....)))))))(((......((((((((((.((((...(((((.......))))).))))))).))))))).......)))... ( -30.02) >DroSec_CAF1 72254 99 + 1 ACCACAGACACGAAGAUAAUUCGUGUGUCUCUCUGUUUUUCUCCCUCGCAUCUGUACCUACGAACGGUAUUUGUGGGGUAGAAGGAACUGCUCGACUAA .......(((((((.....)))))))(((......(((((((.(((((((...(((((.......))))).))))))).))))))).......)))... ( -31.82) >DroSim_CAF1 74065 99 + 1 ACCACAGACACGAAGAUAAUUCGUGUGUCUCUCUCUUUUUCUCCCUCGCAUCUGUACCUACGAACGGUAUUUGUGGGGUAGAAGGAACUGCUCGACUAA ....((((((((((.....))))))).........(((((((.(((((((...(((((.......))))).))))))).))))))).)))......... ( -31.40) >DroEre_CAF1 80437 99 + 1 ACCACAGACACGAAGAUAAUUCGUGUAUAUCUCUCUUUUUCUCCCUCGCAUUCAUACUAACGAACGGUAUUUGCAGGGUAGAAUGAUUUACUCGACUUA .......(((((((.....))))))).......((...((((((((.(((...(((((.......))))).))))))).)))).))............. ( -23.10) >DroYak_CAF1 75527 99 + 1 ACCACAGACACGAAGAUUAUUCGUGUGUCUCUCUCUUUUUCUCCCUCGCAUCUGUACUUACGAACGGAAUUUGUAGGGUAGAAGGACUUACUCGACUUA .......(((((((.....)))))))(((......(((((((((((.((((((((........)))))...))))))).))))))).......)))... ( -25.82) >consensus ACCACAGACACGAAGAUAAUUCGUGUGUCUCUCUCUUUUUCUCCCUCGCAUCUGUACCUACGAACGGUAUUUGUGGGGUAGAAGGAAUUGCUCGACUUA .......(((((((.....))))))).........(((((((.(((((((...(((((.......))))).))))))).)))))))............. (-25.24 = -25.52 + 0.28)

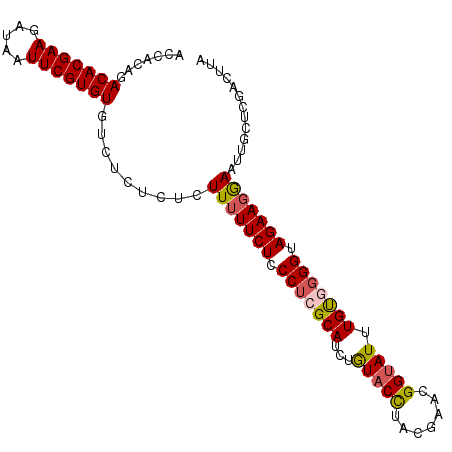
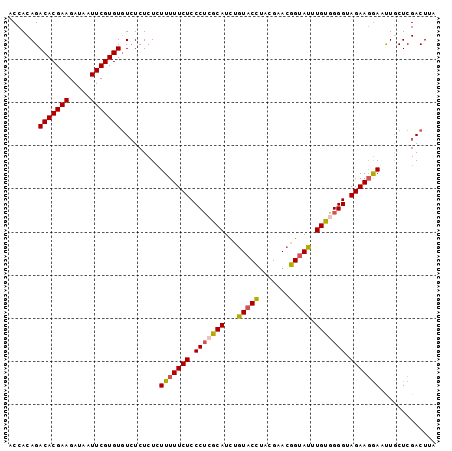
| Location | 22,624,099 – 22,624,206 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -21.72 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22624099 107 + 27905053 CUCUCUCUUUUUCUCCCACGCAUCUGUACCUUCGAACGGUAUUUGUGGGGUAGAAGAAAUUGCUCGACUUAUUAUAUCGUUUGCCACUCAGAAGUUAGGGAAAGAAU---- ........((((.((((..((.((((......((((((((((..((.((((((......)))))).)).....)))))))))).....)))).))..)))).)))).---- ( -29.20) >DroSec_CAF1 72282 102 + 1 CUCUCUGUUUUUCUCCCUCGCAUCUGUACCUACGAACGGUAUUUGUGGGGUAGAAGGAACUGCUCGACUAAAUGUGUCGCUUGUCACACAUAA-----AGGGAGAAU---- ((((((.(((((((.(((((((...(((((.......))))).))))))).))))))).............((((((........))))))..-----))))))...---- ( -33.00) >DroSim_CAF1 74093 107 + 1 CUCUCUCUUUUUCUCCCUCGCAUCUGUACCUACGAACGGUAUUUGUGGGGUAGAAGGAACUGCUCGACUAAAUAUGUCGUUUGUCACACAUAAGUCAAGGAGAGAAU---- .(((((((((((((.(((((((...(((((.......))))).))))))).)))))))....((.((((...(((((.((.....))))))))))).))))))))..---- ( -36.10) >DroEre_CAF1 80465 111 + 1 AUCUCUCUUUUUCUCCCUCGCAUUCAUACUAACGAACGGUAUUUGCAGGGUAGAAUGAUUUACUCGACUUAAUAUGCCGCCUGCCACACAAUAGUUAAAGGCAUAUUUUAU .....((...((((((((.(((...(((((.......))))).))))))).)))).))............((((((((....((.........))....)))))))).... ( -23.70) >DroYak_CAF1 75555 107 + 1 CUCUCUCUUUUUCUCCCUCGCAUCUGUACUUACGAACGGAAUUUGUAGGGUAGAAGGACUUACUCGACUUAAUACGUCACUUGUCACACAGAUGUUAAAGCGAGAUA---- .......(((((((((((.((((((((........)))))...))))))).)))))))....((((.(((...(((((............)))))..)))))))...---- ( -26.10) >consensus CUCUCUCUUUUUCUCCCUCGCAUCUGUACCUACGAACGGUAUUUGUGGGGUAGAAGGAAUUGCUCGACUUAAUAUGUCGCUUGUCACACAGAAGUUAAAGAGAGAAU____ ((((((.(((((((.(((((((...(((((.......))))).))))))).)))))))......((((.......))))...................))))))....... (-21.72 = -22.48 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:25 2006