
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,606,147 – 22,606,277 |
| Length | 130 |
| Max. P | 0.998223 |

| Location | 22,606,147 – 22,606,242 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.16 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22606147 95 + 27905053 AAAUAUCGUGUGCAAAA---------ACAAACCACUUCGUUUCCCAUCACUGAACUUUAAGGGGUGGCAAUAGCAGA--UGCGAAAGCAAA-A-------------AGCGAAAGCAAAUA .......((.(((....---------..((((......)))).(((((.((........)).))))).....))).)--)((....))...-.-------------.((....))..... ( -20.60) >DroSec_CAF1 54500 95 + 1 AAAUAUCGCGUGCAAAA---------ACAAACCACUUCGUUUCCCAUCACUGAACUUUAAGGGGUGGCAAUGGCAGA--UGCGAAAGCAAA-A-------------AGCGAAAGCAAAUA .......((((......---------)).........((((..(((((.((........)).))))).))))))...--(((....)))..-.-------------.((....))..... ( -21.50) >DroSim_CAF1 56289 108 + 1 AAAUAUCGUGUGCAAAA---------ACAAACCACUUCGUUUCCCAUCACUGAACUUUAAGGGGUGGCAAUGGCAGA--UGCGAAAGCAAA-AAGCGAAAGCAAAAAGCGAAAGCAAAUA ........(((......---------))).....(((((((..(((((.((........)).))))).))))).)).--(((....)))..-..((....)).....((....))..... ( -25.80) >DroEre_CAF1 62426 94 + 1 AAAUAUCGUGUGCAAAA---------ACAAACCACUUCGUUUCCCAUCACUGAACAUUGAGGGGUGGCAAAAGCAAG----------------AGCAACAGCAA-AAGCGAAAGCAAAUA .....((((.(((....---------............((..(((.(((........))).)))..))....((...----------------.))....))).-..))))......... ( -19.00) >DroYak_CAF1 56852 119 + 1 AAAUAUCGUUUGCAAAAACAUGCAAAACAAACCACUUCGUUUCCCAUCACUGAACAUUAAAGGGUGGCAAAAGCAAAAUAGCAAAAGCAAGAAAGCAUAAGCGA-AAGCGAAAGCAAAUA .......((((((......((((...............((((.(((((.((.........)))))))...))))......((....))......))))..((..-..))....)))))). ( -24.50) >consensus AAAUAUCGUGUGCAAAA_________ACAAACCACUUCGUUUCCCAUCACUGAACUUUAAGGGGUGGCAAUAGCAGA__UGCGAAAGCAAA_AAGC___AGC_A_AAGCGAAAGCAAAUA ..........(((...............((((......)))).(((((.((........)).))))).....))).....((....))...................((....))..... (-14.86 = -15.16 + 0.30)



| Location | 22,606,178 – 22,606,277 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22606178 99 + 27905053 UUCCCAUCACUGAACUUUAAGGGGUGGCAAUAGCAGA--UGCGAAAGCAAA-A-------------AGCGAAAGCAAAUACCCAUGAGGUUGGAUUCCGCCUCCUUUAGCUCAGU ........(((((.((..(((((((((((((..((..--(((....)))..-.-------------.((....)).........))..))))....))))).)))).)).))))) ( -33.00) >DroSec_CAF1 54531 99 + 1 UUCCCAUCACUGAACUUUAAGGGGUGGCAAUGGCAGA--UGCGAAAGCAAA-A-------------AGCGAAAGCAAAUACCCAUGAGGUUGGAUUCCGCCUCCUUAAGCUCAGU ........(((((.(((.(((((((((((((..((..--(((....)))..-.-------------.((....)).........))..))))....))))).))))))).))))) ( -33.70) >DroSim_CAF1 56320 112 + 1 UUCCCAUCACUGAACUUUAAGGGGUGGCAAUGGCAGA--UGCGAAAGCAAA-AAGCGAAAGCAAAAAGCGAAAGCAAAUACCCAUGAGGUUGGAUUCCGCCUCCUUAAGCUCAGU ........(((((.(((.(((((((((((((..((..--(((....)))..-..((....)).....((....)).........))..))))....))))).))))))).))))) ( -38.20) >DroEre_CAF1 62457 98 + 1 UUCCCAUCACUGAACAUUGAGGGGUGGCAAAAGCAAG----------------AGCAACAGCAA-AAGCGAAAGCAAAUACCCAUGAGGUUGGAUUUUGCCUCCUUAAGCUCAGU ........(((((...((((((((((((....((...----------------.))....))..-..((....))...)))))..(((((........))))))))))..))))) ( -29.10) >DroYak_CAF1 56892 114 + 1 UUCCCAUCACUGAACAUUAAAGGGUGGCAAAAGCAAAAUAGCAAAAGCAAGAAAGCAUAAGCGA-AAGCGAAAGCAAAUACCCAUGAGGUUGGAUUCCGCCUCCUUAAGCUCAGU ........(((((...((((.(((((((....((......))....((......))....))..-..((....))...)))))..(((((.((...))))))).))))..))))) ( -29.00) >consensus UUCCCAUCACUGAACUUUAAGGGGUGGCAAUAGCAGA__UGCGAAAGCAAA_AAGC___AGC_A_AAGCGAAAGCAAAUACCCAUGAGGUUGGAUUCCGCCUCCUUAAGCUCAGU ........(((((.(.((((((((((((....))......((....))...................((....))...)))))..(((((........))))))))))).))))) (-26.98 = -27.32 + 0.34)



| Location | 22,606,178 – 22,606,277 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.14 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22606178 99 - 27905053 ACUGAGCUAAAGGAGGCGGAAUCCAACCUCAUGGGUAUUUGCUUUCGCU-------------U-UUUGCUUUCGCA--UCUGCUAUUGCCACCCCUUAAAGUUCAGUGAUGGGAA ((((((((...((((((((((((((......))))).)))))))))((.-------------.-..(((....)))--...))................))))))))........ ( -28.60) >DroSec_CAF1 54531 99 - 1 ACUGAGCUUAAGGAGGCGGAAUCCAACCUCAUGGGUAUUUGCUUUCGCU-------------U-UUUGCUUUCGCA--UCUGCCAUUGCCACCCCUUAAAGUUCAGUGAUGGGAA (((((((((((((.((.((....((......))((((..(((....((.-------------.-...))....)))--..))))....)).)))))).)))))))))........ ( -30.80) >DroSim_CAF1 56320 112 - 1 ACUGAGCUUAAGGAGGCGGAAUCCAACCUCAUGGGUAUUUGCUUUCGCUUUUUGCUUUCGCUU-UUUGCUUUCGCA--UCUGCCAUUGCCACCCCUUAAAGUUCAGUGAUGGGAA (((((((((((((.((.((....((......))((((..(((....((.....))....((..-...))....)))--..))))....)).)))))).)))))))))........ ( -32.80) >DroEre_CAF1 62457 98 - 1 ACUGAGCUUAAGGAGGCAAAAUCCAACCUCAUGGGUAUUUGCUUUCGCUU-UUGCUGUUGCU----------------CUUGCUUUUGCCACCCCUCAAUGUUCAGUGAUGGGAA ...((((....((((((((((((((......))))).)))))))))((..-..))....)))----------------)..((....))..(((.(((........))).))).. ( -28.90) >DroYak_CAF1 56892 114 - 1 ACUGAGCUUAAGGAGGCGGAAUCCAACCUCAUGGGUAUUUGCUUUCGCUU-UCGCUUAUGCUUUCUUGCUUUUGCUAUUUUGCUUUUGCCACCCUUUAAUGUUCAGUGAUGGGAA (((((((((((((((((((((((((......))))).)))))))))....-..((....((......((....))......))....))......)))).)))))))........ ( -29.20) >consensus ACUGAGCUUAAGGAGGCGGAAUCCAACCUCAUGGGUAUUUGCUUUCGCUU_U_GCU___GCUU_UUUGCUUUCGCA__UCUGCUAUUGCCACCCCUUAAAGUUCAGUGAUGGGAA (((((((((((((((((((((((((......))))).))))))).....................................((....))....)))))).)))))))........ (-24.22 = -24.14 + -0.08)

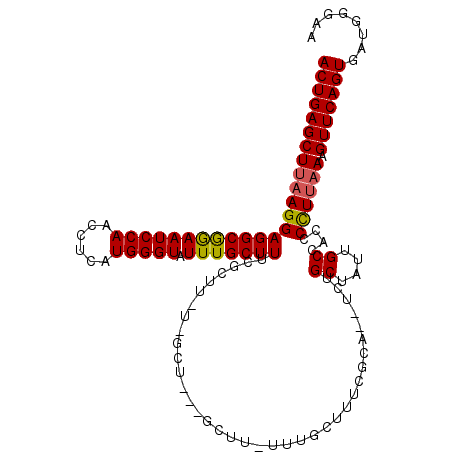

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:10 2006