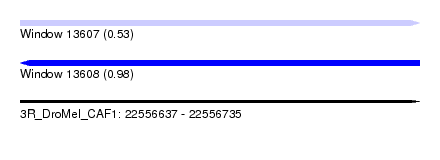
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,556,637 – 22,556,735 |
| Length | 98 |
| Max. P | 0.976857 |

| Location | 22,556,637 – 22,556,735 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22556637 98 + 27905053 UUCAUUUUGU---------UUUUUGUU--UGAAGCCCCACAAUUUGAUAUACACAAAUCGAUUUACUUGCGCAUGCAAAUAACUUUAUUUGCUUAGUGGGUGUUUGGA--A ..........---------......((--(.(((((((((...(((((........))))).............(((((((....)))))))...))))).)))).))--) ( -21.00) >DroVir_CAF1 8499 89 + 1 --------------------CAUUGUUUCUGGAGCGCCACAAUUUGAUACACACAAAUCGAUUUGGUUGCGCAUGUAAAUAACUUUAUUUGCUUAGUGGAAGGAAAAU--A --------------------..((.(((((...((((..(((.(((((........))))).)))...))))..(((((((....))))))).....))))).))...--. ( -20.10) >DroEre_CAF1 4826 91 + 1 UUCAUU-UGU---------UUUUUGUU--UGAAGCGCCACAAUUUGAUAUACACAAAUCGAUUUACUUGCGCAUGCAAAUAACUUUAUUUGCUUAGUGGGC------A--A ......-.((---------(((.....--.)))))(((.....(((((........)))))........(((..(((((((....)))))))...))))))------.--. ( -17.80) >DroWil_CAF1 29031 103 + 1 UAUAUUUUUU------UUUGGGUGGGU--UCGAGUCCCACAAUUUGAUAUACACAAAUCGAUUUACUUGCGCAUGCAAAUAACUUUAUUUGUUUAGUGGGUGGCACACAAA ..........------((((.(((((.--......)))))...(((((........))))).(((((..(....(((((((....)))))))...)..)))))....)))) ( -21.80) >DroAna_CAF1 6031 91 + 1 ----UUCUGU---------UUUUUGUU--UGAAGCCCCACAAUUUGAUAUACACAAAUCGAUUUACUUGCGCAUGCAAAUAACUUUAUUUGCUUAGUGGGUGGAUG----- ----....((---------(((.....--.)))))....(.(((((.......))))).)(((((((..(....(((((((....)))))))...)..))))))).----- ( -18.40) >DroPer_CAF1 9469 107 + 1 UUCGUUUCGCGUUUUUUUUGUACUGUU--UGAAGCCCCACAAUUUGAUAUACACAAAUCGAUUUACUUGCGCAUGCAAAUAACUUUAUUUGCUUAGUGGGUGGCACAA--A ........(((((((............--.)))))(((((...(((((........))))).............(((((((....)))))))...)))))..))....--. ( -19.02) >consensus UUCAUUUUGU_________UUUUUGUU__UGAAGCCCCACAAUUUGAUAUACACAAAUCGAUUUACUUGCGCAUGCAAAUAACUUUAUUUGCUUAGUGGGUGGAACAA__A ...................................(((((...(((((........))))).............(((((((....)))))))...)))))........... (-13.73 = -13.78 + 0.06)

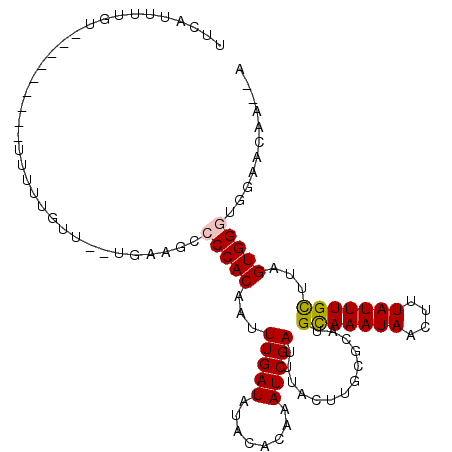
| Location | 22,556,637 – 22,556,735 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22556637 98 - 27905053 U--UCCAAACACCCACUAAGCAAAUAAAGUUAUUUGCAUGCGCAAGUAAAUCGAUUUGUGUAUAUCAAAUUGUGGGGCUUCA--AACAAAAA---------ACAAAAUGAA .--........(((((...(((((((....)))))))((((((((((......))))))))))........)))))......--........---------.......... ( -24.80) >DroVir_CAF1 8499 89 - 1 U--AUUUUCCUUCCACUAAGCAAAUAAAGUUAUUUACAUGCGCAACCAAAUCGAUUUGUGUGUAUCAAAUUGUGGCGCUCCAGAAACAAUG-------------------- .--..((((..........(((..((((....))))..)))((..(((...(((((((.......)))))))))).))....)))).....-------------------- ( -11.90) >DroEre_CAF1 4826 91 - 1 U--U------GCCCACUAAGCAAAUAAAGUUAUUUGCAUGCGCAAGUAAAUCGAUUUGUGUAUAUCAAAUUGUGGCGCUUCA--AACAAAAA---------ACA-AAUGAA .--.------((((((...(((((((....)))))))((((((((((......))))))))))........)))).))....--........---------...-...... ( -23.80) >DroWil_CAF1 29031 103 - 1 UUUGUGUGCCACCCACUAAACAAAUAAAGUUAUUUGCAUGCGCAAGUAAAUCGAUUUGUGUAUAUCAAAUUGUGGGACUCGA--ACCCACCCAAA------AAAAAAUAUA ((((.((.((((........((((((....)))))).((((((((((......))))))))))........)))).)).)))--)..........------.......... ( -22.00) >DroAna_CAF1 6031 91 - 1 -----CAUCCACCCACUAAGCAAAUAAAGUUAUUUGCAUGCGCAAGUAAAUCGAUUUGUGUAUAUCAAAUUGUGGGGCUUCA--AACAAAAA---------ACAGAA---- -----......(((((...(((((((....)))))))((((((((((......))))))))))........)))))......--........---------......---- ( -24.80) >DroPer_CAF1 9469 107 - 1 U--UUGUGCCACCCACUAAGCAAAUAAAGUUAUUUGCAUGCGCAAGUAAAUCGAUUUGUGUAUAUCAAAUUGUGGGGCUUCA--AACAGUACAAAAAAAACGCGAAACGAA (--(((((...(((((...(((((((....)))))))((((((((((......))))))))))........)))))(((...--...)))..........))))))..... ( -28.80) >consensus U__UUCUGCCACCCACUAAGCAAAUAAAGUUAUUUGCAUGCGCAAGUAAAUCGAUUUGUGUAUAUCAAAUUGUGGGGCUUCA__AACAAAAA_________ACAAAAUGAA ...........(((((...(((((((....)))))))((((((((((......))))))))))........)))))................................... (-19.56 = -20.42 + 0.86)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:49 2006