
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,484,126 – 22,484,257 |
| Length | 131 |
| Max. P | 0.920261 |

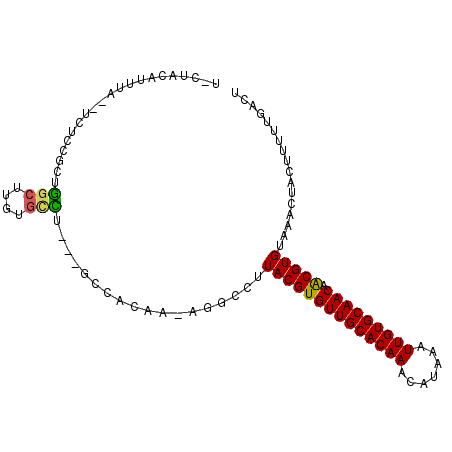
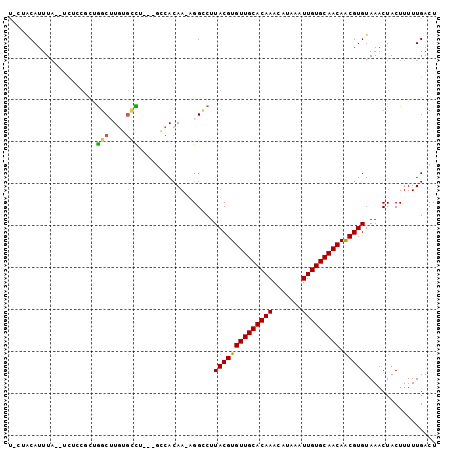
| Location | 22,484,126 – 22,484,225 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -17.97 |
| Energy contribution | -17.63 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
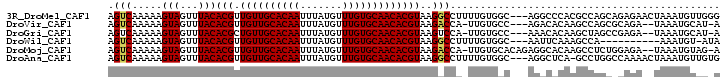
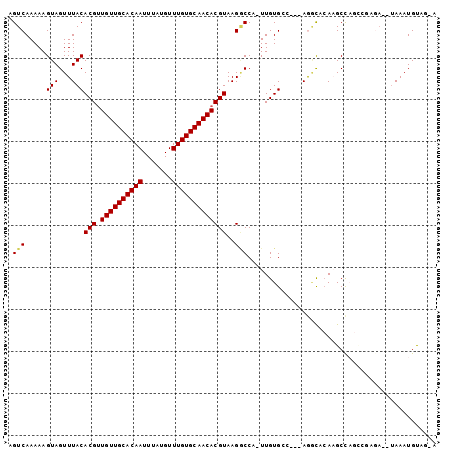
>3R_DroMel_CAF1 22484126 99 + 27905053 CCCAACAUUUAGUUCUCUGCUGGCGUGGGCCU---GCCACAAAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACU ..(((.(..(((((..(.((..((((((((((---........))))).)))))(((((((((.......)))))))))...)))..)))))..).)))... ( -31.00) >DroVir_CAF1 190292 95 + 1 U-AUGCAUUUA--UCUGCGCUGGCUUGUGUCU---GGCACAA-UGGUCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACU .-..(((....--..)))((..((....).).---.))....-((((..((((((((((((((.......))))))))).)))))...)))).......... ( -26.70) >DroGri_CAF1 161896 95 + 1 U-AUGCAUUUA--UCUCGGCUAGCUUGUGUUU---GGCACAA-UGGACUUACGUGUUGCACAAACAUAAAUUGUGCAACAGCGUGUAAACUACUUUUUGACU (-((((.....--...((...((((((((...---..)))).-.)).))..))((((((((((.......)))))))))))))))................. ( -22.40) >DroWil_CAF1 126552 88 + 1 UAU-ACAUUU----------UGGCUUUGAAUU---GCCACAAAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACU ...-......----------((((........---))))(((((((...((((((((((((((.......))))))))).))))).......)))))))... ( -26.40) >DroMoj_CAF1 214007 98 + 1 U-CUACAUUUA--UCUCCAGAGGCUUGUGCCUCUGUGCACAA-UGGUCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACU .-.........--....(((((((....))))))).......-((((..((((((((((((((.......))))))))).)))))...)))).......... ( -31.70) >DroAna_CAF1 128331 98 + 1 CACAACAUUUAGUUUUGGCCAGGC-UGAGCCU---GCCACAAAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACU ...............((((.((((-...))))---))))(((((((...((((((((((((((.......))))))))).))))).......)))))))... ( -31.40) >consensus U_CUACAUUUA__UCUCCGCUGGCUUGUGCCU___GCCACAA_AGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACU .....................(((....)))..................((((((((((((((.......))))))))).)))))................. (-17.97 = -17.63 + -0.33)

| Location | 22,484,126 – 22,484,225 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -17.73 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
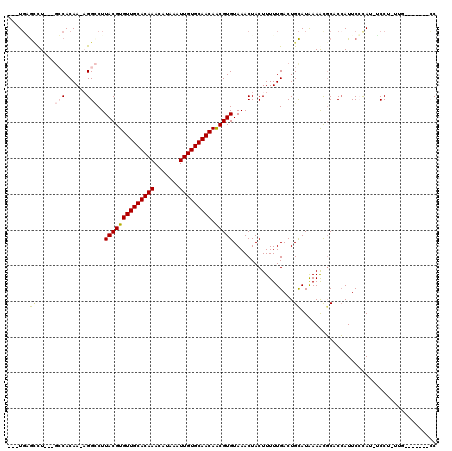
>3R_DroMel_CAF1 22484126 99 - 27905053 AGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUUUGUGGC---AGGCCCACGCCAGCAGAGAACUAAAUGUUGGG ..((((....((((((.(((((.(((((((((.......)))))))))))))...(((......)))---.(((....)))....).))))))....)))). ( -32.70) >DroVir_CAF1 190292 95 - 1 AGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGACCA-UUGUGCC---AGACACAAGCCAGCGCAGA--UAAAUGCAU-A .(((.....(((...)))((((.(((((((((.......)))))))))))))..)))..-(((((..---...)))))......(((..--....)))..-. ( -25.40) >DroGri_CAF1 161896 95 - 1 AGUCAAAAAGUAGUUUACACGCUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGUCCA-UUGUGCC---AAACACAAGCUAGCCGAGA--UAAAUGCAU-A ..........((((((..(((.((((((((((.......))))))))))))).......-..(((..---...))))))))).......--.........-. ( -22.00) >DroWil_CAF1 126552 88 - 1 AGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUUUGUGGC---AAUUCAAAGCCA----------AAAUGU-AUA .....(((((..((((..((((.(((((((((.......))))))))))))).))))))))).((((---........))))----------......-... ( -23.80) >DroMoj_CAF1 214007 98 - 1 AGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGACCA-UUGUGCACAGAGGCACAAGCCUCUGGAGA--UAAAUGUAG-A .(((.....(((...)))((((.(((((((((.......)))))))))))))..)))((-((...(.(((((((....)))))))..).--..))))...-. ( -33.80) >DroAna_CAF1 128331 98 - 1 AGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUUUGUGGC---AGGCUCA-GCCUGGCCAAAACUAAAUGUUGUG ...(((....((((((..((((.(((((((((.......)))))))))))))..((((......(((---.......-))).)))).))))))....))).. ( -33.10) >consensus AGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCA_UUGUGCC___AGGCACAAGCCAGCCGAGA__UAAAUGUAG_A .(((.....(((...)))(((.((((((((((.......)))))))))))))..)))............................................. (-17.73 = -17.73 + -0.00)
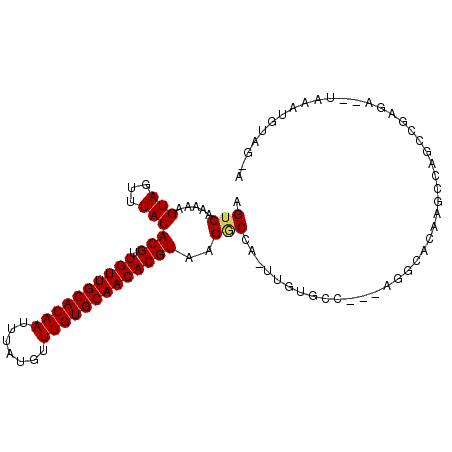
| Location | 22,484,151 – 22,484,257 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
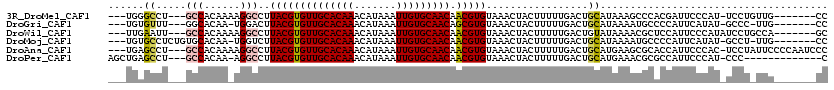
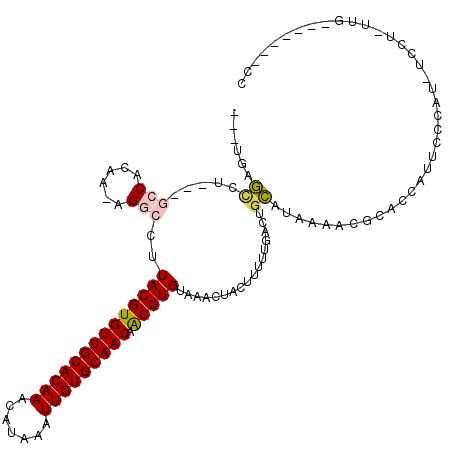
>3R_DroMel_CAF1 22484151 106 + 27905053 ---UGGGCCU---GCCACAAAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCCAU-UCCUGUUG-------CC ---(((((.(---((..(((((((...((((((((((((((.......))))))))).))))).......)))))))...)))....)))))..........-........-------.. ( -30.70) >DroGri_CAF1 161918 104 + 1 ---UGUGUUU---GGCACAA-UGGACUUACGUGUUGCACAAACAUAAAUUGUGCAACAGCGUGUAAACUACUUUUUGACUGCAUAAAAUGCCCCAUUCAUAU-GCCC-UUG-------CC ---...((..---((((.((-(((...((((((((((((((.......)))))))))).)))).................((((...)))).)))))....)-))).-..)-------). ( -27.20) >DroWil_CAF1 126566 107 + 1 ---UUGAAUU---GCCACAAAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGUAUAAAACGCUCCAUUCCCAUAUCCUGCCA-------GC ---..((((.---((..(((((((...((((((((((((((.......))))))))).))))).......)))))))...((.....))))...)))).............-------.. ( -22.20) >DroMoj_CAF1 214029 107 + 1 ---UGUGCCUCUGUGCACAA-UGGUCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAAUGCCCCAUUCAUAU-GCCU-UUG-------CC ---(((((......))))).-((((..((((((((((((((.......))))))))).)))))...))))..........(((((.((((...))))..)))-))..-...-------.. ( -28.00) >DroAna_CAF1 128355 113 + 1 ---UGAGCCU---GCCACAAAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAGCGCACCAUUCCCAC-UCCUAUUCCCCAAUCCC ---((.((.(---((..(((((((...((((((((((((((.......))))))))).))))).......)))))))...)))....)).))..........-................. ( -24.20) >DroPer_CAF1 142102 102 + 1 AGCUGAGCCU---GCCACAA-AGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAACGCGCCAUUCCCAU-CCC-------------C .((...((((---.......-))))......((((((((((.......)))))))))).((((((..(........)..))))))....))...........-...-------------. ( -24.40) >consensus ___UGAGCCU___GCCACAA_AGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAACGCACCAUUCCCAU_UCCU_UUG_______CC ......((.....(((......)))..((((((((((((((.......))))))))).))))).................))...................................... (-17.44 = -17.75 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:19 2006