| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,879,561 – 2,879,654 |
| Length | 93 |
| Max. P | 0.942562 |

| Location | 2,879,561 – 2,879,654 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.34 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -18.49 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
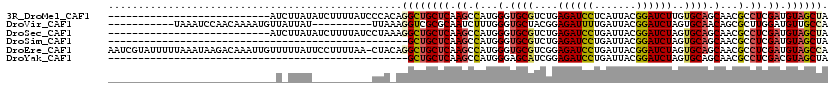
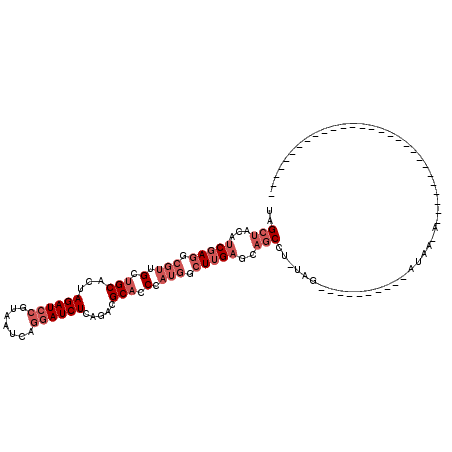
>3R_DroMel_CAF1 2879561 93 + 27905053 ---------------------------AUCUUAUAUCUUUUAUCCCACAGGCUGCUCAAGCCAUGGGUGCGUCUGAGAUCCUCAUUACGGAUCUUGUGCAGCAACGCCUCGAUGUAGCUA ---------------------------...(((((((......((((..((((.....)))).))))(((((..(((((((.......)))))))..)).))).......)))))))... ( -29.20) >DroVir_CAF1 729 99 + 1 -----------UAAAUCCAACAAAAUGUUAUUAU----------UUAAAGGUCGCGCAAUCUUUGGGUGCUACGGAGAUUUUGAUUACGGAUCUAGUGCAACAGCGCUUGGAUGUUGCCA -----------...((((((((((((.((..(((----------((((((((......))))))))))).....)).))))))............((((....))))))))))....... ( -21.60) >DroSec_CAF1 1243 93 + 1 ---------------------------AUCUUAUAUCUUUUAUCCUAAAGGCUGCUCAAGCCAUGGGUGCGUCUGAGAUCCUGAUUACGGAUCUAGUGCAGCAACGCCUCGAUGUAGCUA ---------------------------...(((((((............((((.....))))..(((.(((((((((((((.......))))))....)))..))))))))))))))... ( -27.70) >DroSim_CAF1 1 70 + 1 --------------------------------------------------GCUGCUCAAGCCAUGGGUGCGUCUGAGAUCCUGAUUACGGAUCUAGUGCAGCAACGCCUCGAUGUAGCUA --------------------------------------------------(((((((.((.(...(.((((.((.((((((.......)))))))))))).)...).)).)).))))).. ( -23.60) >DroEre_CAF1 1208 119 + 1 AAUCGUAUUUUUAAAUAAGACAAAUUGUUUUUAUUCCUUUUAA-CUACAGGCUGCUCAAGCCAUGGGUGCGUCGGAGAUCCUGAUUACGGAUCUAGUGCAGCAACGCCUCGAUGUAGCCA ....(((...(((((.(((((.....))))).......)))))-.))).((((.....)))).(((.((((((((((((((.......)))))).(((......))).)))))))).))) ( -30.40) >DroYak_CAF1 1 70 + 1 --------------------------------------------------GCUGCUCAAGCCAUGGGAGCAUCGGAGAUCCUGAUUACGGAUCUAGUGCAGCAACGCCUCGACGUAGCUA --------------------------------------------------(((((.(...(((((....))).))((((((.......)))))).).))))).(((......)))..... ( -21.90) >consensus ____________________________U_UUAU__________CUA_AGGCUGCUCAAGCCAUGGGUGCGUCGGAGAUCCUGAUUACGGAUCUAGUGCAGCAACGCCUCGAUGUAGCUA .................................................((((((((.((.(...(.((((....((((((.......))))))..)))).)...).)).)).)))))). (-18.49 = -18.88 + 0.39)
| Location | 2,879,561 – 2,879,654 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.34 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -15.60 |
| Energy contribution | -17.43 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2879561 93 - 27905053 UAGCUACAUCGAGGCGUUGCUGCACAAGAUCCGUAAUGAGGAUCUCAGACGCACCCAUGGCUUGAGCAGCCUGUGGGAUAAAAGAUAUAAGAU--------------------------- .......(((...((((..(((....((((((.......))))))))))))).(((((((((.....)))).)))))......))).......--------------------------- ( -28.00) >DroVir_CAF1 729 99 - 1 UGGCAACAUCCAAGCGCUGUUGCACUAGAUCCGUAAUCAAAAUCUCCGUAGCACCCAAAGAUUGCGCGACCUUUAA----------AUAAUAACAUUUUGUUGGAUUUA----------- ..((((((.(.....).))))))..(((((((((((((.....................))))))..........(----------((((.......))))))))))))----------- ( -17.30) >DroSec_CAF1 1243 93 - 1 UAGCUACAUCGAGGCGUUGCUGCACUAGAUCCGUAAUCAGGAUCUCAGACGCACCCAUGGCUUGAGCAGCCUUUAGGAUAAAAGAUAUAAGAU--------------------------- ..(((...(((((.(((.(.(((.((((((((.......)))))).))..))).).))).)))))..)))((((......)))).........--------------------------- ( -24.70) >DroSim_CAF1 1 70 - 1 UAGCUACAUCGAGGCGUUGCUGCACUAGAUCCGUAAUCAGGAUCUCAGACGCACCCAUGGCUUGAGCAGC-------------------------------------------------- ..(((...(((((.(((.(.(((.((((((((.......)))))).))..))).).))).)))))..)))-------------------------------------------------- ( -23.50) >DroEre_CAF1 1208 119 - 1 UGGCUACAUCGAGGCGUUGCUGCACUAGAUCCGUAAUCAGGAUCUCCGACGCACCCAUGGCUUGAGCAGCCUGUAG-UUAAAAGGAAUAAAAACAAUUUGUCUUAUUUAAAAAUACGAUU .((((...(((((.(((.(.(((...((((((.......)))))).....))).).))).)))))..))))((((.-(((((((((.((((.....)))))))).)))))...))))... ( -31.10) >DroYak_CAF1 1 70 - 1 UAGCUACGUCGAGGCGUUGCUGCACUAGAUCCGUAAUCAGGAUCUCCGAUGCUCCCAUGGCUUGAGCAGC-------------------------------------------------- ..(((.(.(((((.(((.(..(((..((((((.......))))))....)))..).))).)))))).)))-------------------------------------------------- ( -22.30) >consensus UAGCUACAUCGAGGCGUUGCUGCACUAGAUCCGUAAUCAGGAUCUCAGACGCACCCAUGGCUUGAGCAGCCU_UAG__________AUAA_A____________________________ ..(((...(((((.(((.(.(((...((((((.......)))))).....))).).))).)))))..))).................................................. (-15.60 = -17.43 + 1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:34 2006