
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,475,189 – 22,475,305 |
| Length | 116 |
| Max. P | 0.731538 |

| Location | 22,475,189 – 22,475,305 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -28.78 |
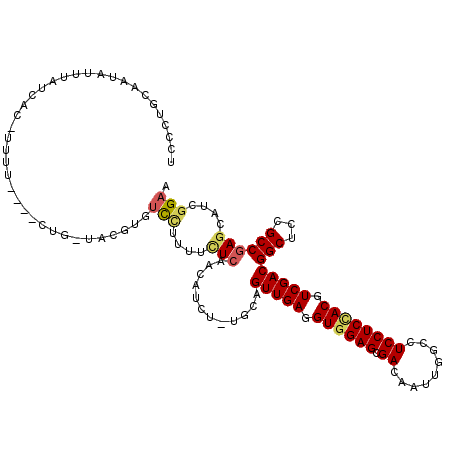
| Consensus MFE | -21.05 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
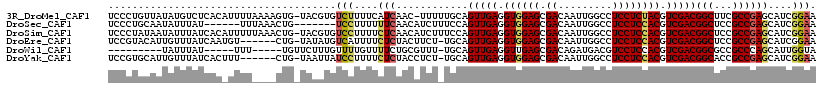
>3R_DroMel_CAF1 22475189 116 - 27905053 UCCCUGUUAUAUGUCUCACAUUUUAAAAGUG-UACGUGUCUUUUCAUCAAC-UUUUUGCAGUUGAGGUGGAGCGACAAUUGGCCUCCUCUACGUCGACGGCUUCGCCGAGCAUCGGAA (((.....((((((..(((.........)))-.))))))............-....(((.(((((.((((((.((.........)))))))).)))))(((...)))..)))..))). ( -28.80) >DroSec_CAF1 108117 105 - 1 UCCCUGCAAUAUUUAU------UUUAAACUG-------UCCUUUUUUCAACAUCUUUCCAGUUGAGGUGGAGCGACAAUUGGCCUCCUCCACGUCGACGGCUCCGCCGAGCAUCGGAA (((.(((.........------.........-------........(((((.........)))))((((((((......(((......)))((....))))))))))..)))..))). ( -27.90) >DroSim_CAF1 105822 117 - 1 UCCCUAUAAUAUUUAUCACAUUUUUAAACUG-UACGUGUCCUUUUCUCAACAUCUUUCCAGUUGAGGUGGAGCGACAAUUGGCCUCCUCCACGUCGACGGCUCCGCCGAGCAUCGGAA ...............................-......(((..(.(((............(((((.((((((.((.........)))))))).)))))(((...)))))).)..))). ( -27.00) >DroEre_CAF1 109169 110 - 1 UCCGUACAUUGUUUAUCAAUGU------CUG-UAUAUGUCAUUUUCUCUACUUCU-UGCAGUUGAGGUGGAGCGACAAUUGGCCUCCUCCACGUCGACGGCUCCGCCGAGCAUCGGAA ((((.((((((.....))))))------...-.......................-(((.(((((.((((((.((.........)))))))).)))))(((...)))..))).)))). ( -32.70) >DroWil_CAF1 117269 98 - 1 ---------UAUUUAU-----UUU-----UGUUCUUUGUUUUGUUUUCUGCGUUU-UGCAGUUGAGGUUGAGCGACAGAUGACGUCCUCCACGUCGACGGCGCCGCCCAGCAUUGGUA ---------.......-----...-----..................(((.((..-(((.(((((.((.(((.(((.......)))))).)).))))).)))..)).)))........ ( -22.20) >DroYak_CAF1 107975 110 - 1 UCCGUGCAUUGUUUAUCACUUU------CUG-UAAUUAUCCUUUUCUCUACCUCU-UGCAGUUGAGGUGGAGCGACAAUUGGCCUCCUCCACGUCGACGGCACCGCCGAGCAUCGGAA (((((((...............------...-.............(((((((((.-.......)))))))))((((...(((......))).)))).((((...)))).))).)))). ( -34.10) >consensus UCCCUGCAAUAUUUAUCAC_UUUU____CUG_UACGUGUCCUUUUCUCAACAUCU_UGCAGUUGAGGUGGAGCGACAAUUGGCCUCCUCCACGUCGACGGCUCCGCCGAGCAUCGGAA ......................................(((....(((............(((((.((((((.((.........)))))))).)))))(((...))))))....))). (-21.05 = -21.30 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:15 2006