
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,460,516 – 22,460,653 |
| Length | 137 |
| Max. P | 0.992196 |

| Location | 22,460,516 – 22,460,613 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -17.23 |
| Energy contribution | -19.29 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
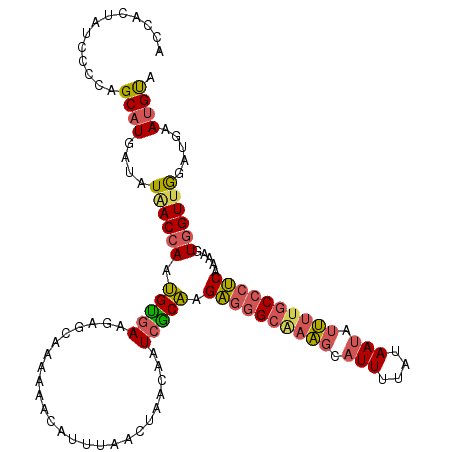
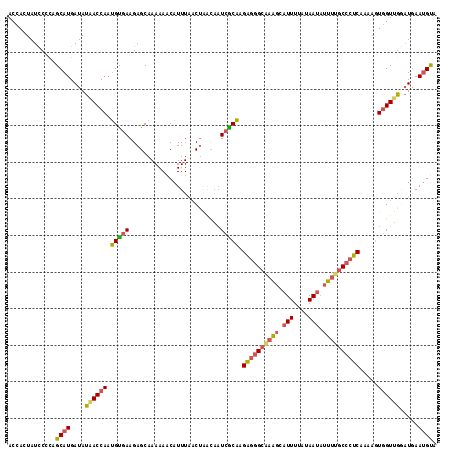
>3R_DroMel_CAF1 22460516 97 - 27905053 AAAACAUUUAACUGACAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAUGUGGUUGGAUGAAUGUA--UCCAAAUCGGGAAUA----------CGGCC ...(((((((.....((((((((.((((((((((.(((....))).))))))))))...))))))))..))))))).--(((......)))...----------..... ( -31.10) >DroGri_CAF1 137981 95 - 1 CGAUCGUUUGUCGGGCAAUCCACAAGCCUUAAAGCGUUUUAUAAUCUUU-GCC-------------AGGGAACUGUAAAACCAAAUAGGGAAUAAUGGUGGAAUAGUCC .(((.....)))((((..(((((...(((((....((((((((.((((.-...-------------.))))..))))))))....))))).......)))))...)))) ( -21.70) >DroSec_CAF1 93756 97 - 1 AAAACAUUUAACUAGCAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAAGUGGUUGGAUGAAUGUA--UCCAAAUCGGGAAUA----------CGGCC ....((((((((((((....))..((((((((((.(((....))).)))))))))).....))))))))))..((((--(((......)).)))----------))... ( -31.80) >DroSim_CAF1 90522 97 - 1 AAAACAUUUAACUAACAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAAGUGGUUAGAUGAAUGUA--UCCAAAUCGGGAAUA----------CGGCC ...(((((((.(((((...(....((((((((((.(((....))).))))))))))....)..))))).))))))).--(((......)))...----------..... ( -29.60) >DroEre_CAF1 94433 97 - 1 AAAGCAUUUAACUAAAAAUCGCAAGAGGGCUAAGCAUUUUAUAAUAUUUUGCCCUCAAAAAUGGUUGCAUGAAUGUA--UCCAAAUCGGGCAUA----------CUGCC ...((((((......)))).))..((((((.(((.(((....))).))).)))))).....(((.((((....))))--.))).....(((...----------..))) ( -22.10) >DroYak_CAF1 93283 97 - 1 AAAACAUUUAACUCACAAUCGCAAGAGGGCAAAGCAUUUUAUAACAUUUUGCCCUCAAAAGUGGUUGGAUGAAUGUA--ACCAAAUCGGGAAUA----------CUGCC ...(((((((..((.(((((((..((((((((((..((....))..))))))))))....)))))))))))))))).--.((.....)).....----------..... ( -27.80) >consensus AAAACAUUUAACUAACAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAAGUGGUUGGAUGAAUGUA__UCCAAAUCGGGAAUA__________CGGCC ...(((((((.(((((........((((((((((.(((....))).)))))))))).......))))).)))))))....((.....)).................... (-17.23 = -19.29 + 2.06)



| Location | 22,460,536 – 22,460,653 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.83 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22460536 117 - 27905053 ACCACUAUCCCCAGCAUGAUAUAACCAAUGUGAAGAGCAAAAAACAUUUAACUGACAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAUGUGGUUGGAUGAAUGUA .......((....((((..........))))...)).......(((((((.....((((((((.((((((((((.(((....))).))))))))))...))))))))..))))))). ( -30.10) >DroSec_CAF1 93776 117 - 1 ACCACUAUCCCUAGCAUGAUAUAACCAAUGUGAAGAGCAAAAAACAUUUAACUAGCAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAAGUGGUUGGAUGAAUGUA .............((((...........(((.....))).....((((((((((((....))..((((((((((.(((....))).)))))))))).....)))))))))).)))). ( -29.80) >DroSim_CAF1 90542 117 - 1 ACCACUAUCCCCAGCAUGAUAUAACCAAUGUGAAGAGCAAAAAACAUUUAACUAACAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAAGUGGUUAGAUGAAUGUA .......((....((((..........))))...)).......(((((((.(((((...(....((((((((((.(((....))).))))))))))....)..))))).))))))). ( -28.60) >DroEre_CAF1 94453 117 - 1 ACCACUAUUCCCAGCAUGAUAUGACCAAUGCGAAGAGCAAAAAGCAUUUAACUAAAAAUCGCAAGAGGGCUAAGCAUUUUAUAAUAUUUUGCCCUCAAAAAUGGUUGCAUGAAUGUA .............((((.((..(((((.(((((...((.....)).(((....)))..))))).((((((.(((.(((....))).))).)))))).....)))))..))..)))). ( -26.40) >DroYak_CAF1 93303 117 - 1 ACCACUAUUCCCAGCAUGAUAUAACCAAUGCGAAGAGCAAAAAACAUUUAACUCACAAUCGCAAGAGGGCAAAGCAUUUUAUAACAUUUUGCCCUCAAAAGUGGUUGGAUGAAUGUA .......(((...((((..........))))...)))......(((((((..((.(((((((..((((((((((..((....))..))))))))))....)))))))))))))))). ( -30.70) >DroAna_CAF1 104577 117 - 1 ACCAGUAUUCCCAGCAUGAUAUAACCACGGUGAAGAGCAAAAAACAUUUGUCAGCUCCUAACCAGUCUGCGAGCCAUUUUAUAAUUUCAUUCCCGCAAAAGUCGUCGACUAAAAGCA .............((.............(((...((((...............))))...)))((((.((((.............................)))).))))....)). ( -16.91) >consensus ACCACUAUCCCCAGCAUGAUAUAACCAAUGUGAAGAGCAAAAAACAUUUAACUAACAAUCGCAAGAGGGCAAAGCAUUUUAUAAUAUUUUGCCCUCAAAAGUGGUUGGAUGAAUGUA .............((((....((((((.(((((.........................))))).((((((((((.(((....))).)))))))))).....)))))).....)))). (-22.68 = -22.83 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:13 2006