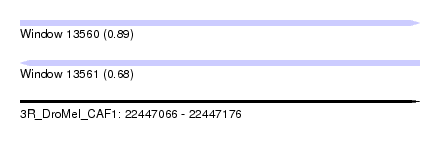
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,447,066 – 22,447,176 |
| Length | 110 |
| Max. P | 0.892199 |

| Location | 22,447,066 – 22,447,176 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.41 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -14.99 |
| Energy contribution | -14.93 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22447066 110 + 27905053 AUAUUCUAGACGAAUGAAUAAUAUAGCAUUCAUUAUC-UA-CCUUUUAUACCGCCAACGCGAC-GACGUUUAAAAUUAGACUAGAAAAUCUAGUC-AAU-UGAGCCGUCUAGUCA-- .....((((((((((((((........)))))))...-..-..........(((....)))..-...((((((.....((((((.....))))))-..)-))))))))))))...-- ( -25.50) >DroVir_CAF1 144425 113 + 1 -UGUUCUUUUAGAAUGCUAGCCAUGUC-UAUAUAAACUUA-GCUUUUAUACCGCCAACGCGGCCAGCGACUAUAAUUAGACCAGUUGAUCUAGCCGCAU-UAAACCGUCGCGCUCUC -.((((.....))))(((((.((.(((-((..........-(((......((((....))))..))).........)))))....))..)))))(((..-.........)))..... ( -19.21) >DroPse_CAF1 101706 110 + 1 CUGUUCUAAUAGAAUGCGGGAUGUAUC-UUUAUUAAC-UA-GCUUUUAUACCGCCAACGCGAA-GACGGUUAAAAUUAGACCAGAAAAUCUGGUCGAAU-UGAGCCGUCGCAUAG-- ......(((((((((((.....)))).-))))))).(-((-((........(((....)))..-(((((((..((((.((((((.....)))))).)))-).))))))))).)))-- ( -31.60) >DroWil_CAF1 86938 108 + 1 CGAUUCUAUUAGAAUGAC-AACAAGUUGCUCAUUAGC-AAUGCUUUUAUACCGCCAACGCGAU-GGCUGCUAAAAUUAGACCAGUCUAUCUGGUCGAAU-UAAACUGUCGCA----- ..((((.....))))(((-(....((((((....)))-)))...........((((......)-)))......((((.((((((.....)))))).)))-)....))))...----- ( -25.20) >DroAna_CAF1 90633 110 + 1 GUGUUCUAUAAGAAUGCUGCCUGUAUC-UUUAUUAAC-UA-GCUUUUAUACCGCCAACGCGAC-GACGCUUUAAAUUAGACUAGAAAAUCUAGUC-GAUUUGAGCCGUCGAAUCG-- (((((.(((((((..((((...(((..-..)))....-))-))))))))).....)))))..(-((((.((((((((.((((((.....))))))-)))))))).))))).....-- ( -29.00) >DroPer_CAF1 102384 110 + 1 CUGUUCUAAUAGAAUGCGGGAUGUAUC-UUUAUUAAC-UA-GCUUUUAUACCGCCAACGCGAA-GACGGUUAAAAUUAGACCAGAAAAUCUGGUCGAAU-UGAGCCGUCGCAUAG-- ......(((((((((((.....)))).-))))))).(-((-((........(((....)))..-(((((((..((((.((((((.....)))))).)))-).))))))))).)))-- ( -31.60) >consensus CUGUUCUAUUAGAAUGCGGGAUAUAUC_UUUAUUAAC_UA_GCUUUUAUACCGCCAACGCGAC_GACGGUUAAAAUUAGACCAGAAAAUCUAGUCGAAU_UGAGCCGUCGCAUAG__ ..((((.....))))....................................(((....)))...((((......(((.((((((.....)))))).)))......))))........ (-14.99 = -14.93 + -0.05)

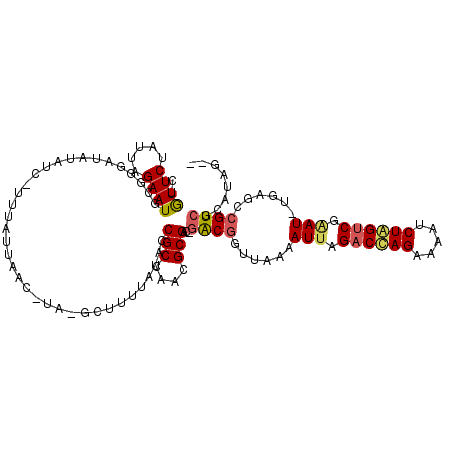
| Location | 22,447,066 – 22,447,176 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.41 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22447066 110 - 27905053 --UGACUAGACGGCUCA-AUU-GACUAGAUUUUCUAGUCUAAUUUUAAACGUC-GUCGCGUUGGCGGUAUAAAAGG-UA-GAUAAUGAAUGCUAUAUUAUUCAUUCGUCUAGAAUAU --...((((((((((..-...-((((((.....))))))...(((((..((((-(......)))))...)))))))-).-...((((((((......)))))))))))))))..... ( -29.50) >DroVir_CAF1 144425 113 - 1 GAGAGCGCGACGGUUUA-AUGCGGCUAGAUCAACUGGUCUAAUUAUAGUCGCUGGCCGCGUUGGCGGUAUAAAAGC-UAAGUUUAUAUA-GACAUGGCUAGCAUUCUAAAAGAACA- .((((.((.((.(((.(-((((((((((....((((.........))))..)))))))))))))).)).....(((-((.((((....)-))).))))).)).)))).........- ( -35.30) >DroPse_CAF1 101706 110 - 1 --CUAUGCGACGGCUCA-AUUCGACCAGAUUUUCUGGUCUAAUUUUAACCGUC-UUCGCGUUGGCGGUAUAAAAGC-UA-GUUAAUAAA-GAUACAUCCCGCAUUCUAUUAGAACAG --..(((((((((...(-(((.((((((.....)))))).))))....)))))-.((..(((((((((......))-).-))))))...-))........))))............. ( -27.00) >DroWil_CAF1 86938 108 - 1 -----UGCGACAGUUUA-AUUCGACCAGAUAGACUGGUCUAAUUUUAGCAGCC-AUCGCGUUGGCGGUAUAAAAGCAUU-GCUAAUGAGCAACUUGUU-GUCAUUCUAAUAGAAUCG -----.(((((((((..-.(((((((((.....)))))).....(((((((((-(......)))).((......))..)-))))).))).))).))))-)).((((.....)))).. ( -27.90) >DroAna_CAF1 90633 110 - 1 --CGAUUCGACGGCUCAAAUC-GACUAGAUUUUCUAGUCUAAUUUAAAGCGUC-GUCGCGUUGGCGGUAUAAAAGC-UA-GUUAAUAAA-GAUACAGGCAGCAUUCUUAUAGAACAC --.(.(((((((((.(((((.-((((((.....))))))..))))...).)))-)))..((((.(.((((......-..-.........-.)))).).)))).........)))).. ( -23.47) >DroPer_CAF1 102384 110 - 1 --CUAUGCGACGGCUCA-AUUCGACCAGAUUUUCUGGUCUAAUUUUAACCGUC-UUCGCGUUGGCGGUAUAAAAGC-UA-GUUAAUAAA-GAUACAUCCCGCAUUCUAUUAGAACAG --..(((((((((...(-(((.((((((.....)))))).))))....)))))-.((..(((((((((......))-).-))))))...-))........))))............. ( -27.00) >consensus __CGAUGCGACGGCUCA_AUUCGACCAGAUUUUCUGGUCUAAUUUUAACCGUC_GUCGCGUUGGCGGUAUAAAAGC_UA_GUUAAUAAA_GAUACAUCCAGCAUUCUAAUAGAACAG ........(((((.....(((.((((((.....)))))).))).....)))))..((((....)))).................................................. (-15.22 = -15.42 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:06 2006