
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,436,198 – 22,436,308 |
| Length | 110 |
| Max. P | 0.741386 |

| Location | 22,436,198 – 22,436,308 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.68 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -10.47 |
| Energy contribution | -10.33 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
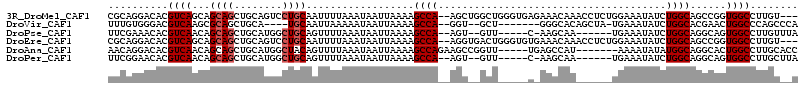
>3R_DroMel_CAF1 22436198 110 + 27905053 ---ACAAGGCCACCGGCUGCCAGAUAUUUCCAGAGGUUUGUUUCUCACCCAGCCAGCU--UGGCUUUUAAUUAUUUAAAAUUGCAGGACUGCAGCUGCUGCUGACGUGUCCUGCG ---..((((((((.(((((...((....))..((((......))))...))))).)..--)))))))...............(((((((((((((....)))).)).))))))). ( -38.60) >DroVir_CAF1 132839 99 + 1 UGGGCUGGGCCAGUUCGUGCCAGAUAUUUCA-UAGCUGUGCCC-------AGC--ACC--UGGCUUUUAAUUAUUUUUAAUUGCA----UGCAGCUGCGCUUGACGUCCCACAAA (((((((((((((((.(((..........))-)))))).))))-------)((--(.(--((((..((((......))))..)).----..))).))).......).)))).... ( -28.60) >DroPse_CAF1 89934 99 + 1 UAAACAAGGCCACUGCCUGCCAGAUAUUUCA------UUGCUU-G-----AAC--ACU--UGGCUUUUAAUUAUUUAAAACUGCAGCCAUGCAGCUGCUGUUGACGUGUUUCGAA .(((((((((....))))(((((....((((------.....)-)-----)).--..)--))))..............(((.(((((......))))).)))....))))).... ( -23.10) >DroEre_CAF1 68126 110 + 1 ---ACAAGGCCACCGGCUGCCAGAUAUUUCCAGAGGUUUGUUUCACACCCAGUCACCU--UGGCUUUUAAUUAUUUAAAAUUGCAGGACUGCAGCUGCUGCUGACGUGUCCUGCG ---.(((((..((.((.((..(((((..((....))..)))))..)).)).))..)))--))..((((((....))))))..(((((((((((((....)))).)).))))))). ( -34.20) >DroAna_CAF1 75708 103 + 1 GGUGCAAGGCCAGUGCCUGCCAUAUAUUUU-------AUGGCUCA-----AACCGGCUUCUGGCUUUUAAUUAUUUAAAACUGUAGCCAUGCAGCUGCUGUUGACGUGUCCUGUU ((..(((((((((.(((.((((((.....)-------)))))...-----....)))..))))))))...........(((.(((((......))))).)))...)..))..... ( -33.00) >DroPer_CAF1 90865 99 + 1 UAAGCAAGGCCACUGCCUGCCAGAUAUUUCA------UUGCUU-G-----AAC--ACU--UGGCUUUUAAUUAUUUAAAACUGCAGCCAUGCAGCUGCUGUUGACGUGUUCCGAA .(((((((((........))).((....)).------))))))-(-----(((--(((--..((((((((....))))))..(((((......))))).))..).)))))).... ( -28.60) >consensus U__ACAAGGCCACUGCCUGCCAGAUAUUUCA______UUGCUU_A_____AAC__ACU__UGGCUUUUAAUUAUUUAAAACUGCAGCCAUGCAGCUGCUGUUGACGUGUCCCGAA ...((..(((........)))........................................(((((((((....))))))(((((....)))))..)))......))........ (-10.47 = -10.33 + -0.14)



| Location | 22,436,198 – 22,436,308 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.68 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -9.76 |
| Energy contribution | -10.09 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22436198 110 - 27905053 CGCAGGACACGUCAGCAGCAGCUGCAGUCCUGCAAUUUUAAAUAAUUAAAAGCCA--AGCUGGCUGGGUGAGAAACAAACCUCUGGAAAUAUCUGGCAGCCGGUGGCCUUGU--- .(((((((..(.((((....))))).)))))))..((((((....))))))(((.--.(((((((((((.........))).(..((....))..)))))))))))).....--- ( -40.30) >DroVir_CAF1 132839 99 - 1 UUUGUGGGACGUCAAGCGCAGCUGCA----UGCAAUUAAAAAUAAUUAAAAGCCA--GGU--GCU-------GGGCACAGCUA-UGAAAUAUCUGGCACGAACUGGCCCAGCCCA ....((((..((((...(((......----)))..................((((--(((--(((-------(....))))..-......)))))))......))))....)))) ( -26.50) >DroPse_CAF1 89934 99 - 1 UUCGAAACACGUCAACAGCAGCUGCAUGGCUGCAGUUUUAAAUAAUUAAAAGCCA--AGU--GUU-----C-AAGCAA------UGAAAUAUCUGGCAGGCAGUGGCCUUGUUUA ...(((((..(....).((((((....)))))).)))))((((((......((((--..(--(((-----.-.(....------)..))))..)))).(((....))))))))). ( -26.00) >DroEre_CAF1 68126 110 - 1 CGCAGGACACGUCAGCAGCAGCUGCAGUCCUGCAAUUUUAAAUAAUUAAAAGCCA--AGGUGACUGGGUGUGAAACAAACCUCUGGAAAUAUCUGGCAGCCGGUGGCCUUGU--- .(((((((..(.((((....))))).)))))))..((((((....))))))..((--((((.(((((.(((((........)).(((....))).))).))))).)))))).--- ( -39.50) >DroAna_CAF1 75708 103 - 1 AACAGGACACGUCAACAGCAGCUGCAUGGCUACAGUUUUAAAUAAUUAAAAGCCAGAAGCCGGUU-----UGAGCCAU-------AAAAUAUAUGGCAGGCACUGGCCUUGCACC ..((((.....((....((....)).(((((..((((......))))...))))))).((((((.-----...(((((-------(.....))))))....)))))))))).... ( -26.60) >DroPer_CAF1 90865 99 - 1 UUCGGAACACGUCAACAGCAGCUGCAUGGCUGCAGUUUUAAAUAAUUAAAAGCCA--AGU--GUU-----C-AAGCAA------UGAAAUAUCUGGCAGGCAGUGGCCUUGCUUA ....((((((.......((((((....))))))..((((((....))))))....--.))--)))-----)-......------..........(((((((....))).)))).. ( -27.20) >consensus UUCAGGACACGUCAACAGCAGCUGCAUGGCUGCAAUUUUAAAUAAUUAAAAGCCA__AGU__GCU_____C_AAGCAA______UGAAAUAUCUGGCAGGCAGUGGCCUUGC__A ..........((((...((((........))))..................((((......................................))))......))))........ ( -9.76 = -10.09 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:03 2006