
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,435,915 – 22,436,016 |
| Length | 101 |
| Max. P | 0.942313 |

| Location | 22,435,915 – 22,436,016 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -33.59 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
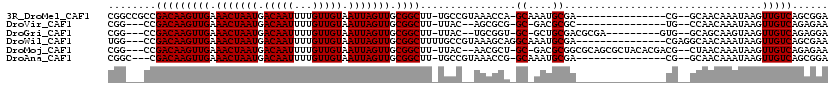
>3R_DroMel_CAF1 22435915 101 + 27905053 CGGCCGCCGACAAGUUGAAACUAAUGACAAUUUUGUUGUAAUUAGUUGCGGCUU-UGCCGUAAACCA-GCAAAUGCGA---------------CG--GCAACAAAUAAGUUGUCAGCGGA ...((((.(((((((((.(((((((.(((((...))))).))))))).)))).(-((((((......-((....)).)---------------))--))))........))))).)))). ( -36.30) >DroVir_CAF1 132509 95 + 1 CGG---CCGACAAGUUGAAACUAAUGACAAUUUUGUUGUAAUUAGUUGCGGCUU-UUAC--AGCGCG-GC-GACGCGC---------------UG--CCAACAAAUAAGUUGUCAGAGAA .((---(.(((((((((.(((((((.(((((...))))).))))))).))))))-...(--((((((-..-..)))))---------------))--...........))))))...... ( -32.80) >DroGri_CAF1 110574 101 + 1 CGG---CCGACAAGUUGAAACUAAUGACAAUUUUGUUGUAAUUAGUUGCGGCUU-UUAC--UGCGGU-GC-GCUGCGACGCGA---------GUG--GCAGCAAGUAAGUUGUCAGAGGA ...---..(((((((((.(((((((.(((((...))))).))))))).)))).(-((((--(((...-))-(((((.((....---------)).--))))).)))))))))))...... ( -35.40) >DroWil_CAF1 73644 102 + 1 UGG---CCGACAAGUUGAAACUAAUGACAAUUUUGUUGUAAUUAGUUGCGGCUUUUGCCGUAAAGCAGGCAAAUGCGA---------------CGAGGCAACAAAUAAGUUGUCAGCGAA ..(---((((....))).......(((((((((((((((.....((((((...((((((........)))))))))))---------------)...))))))))...)))))))))... ( -31.50) >DroMoj_CAF1 161120 110 + 1 CGG---CCGACAAGUUGAAACUAAUGACAAUUUUGUUGUAAUUAGUUGCGGCUU-UUAC--AACGCU-GC-GACGCGGCGCAGCGCUACACGACG--CUAACAAAUAAGUUGUCAGAGAA ...---..(((((.(((.(((((((.(((((...))))).)))))))(((....-....--..((((-((-(......)))))))........))--).......))).)))))...... ( -31.27) >DroAna_CAF1 75423 98 + 1 CGGC---CGACAAGUUGAAACUAAUGACAAUUUUGUUGUAAUUAGUUGCGGCUU-UGCCGUAAACCG-GCAAAUGCGA---------------CG--GCAACAAAUAAGUUGUCAGCGGA ...(---((.(.(((....)))..(((((((((((((((.....((((((..((-(((((.....))-))))))))))---------------).--))))))))...))))))))))). ( -34.30) >consensus CGG___CCGACAAGUUGAAACUAAUGACAAUUUUGUUGUAAUUAGUUGCGGCUU_UGAC__AAAGCA_GC_AAUGCGA_______________CG__GCAACAAAUAAGUUGUCAGAGAA ........(((((((((.(((((((.(((((...))))).))))))).))))................((....)).................................)))))...... (-17.80 = -17.80 + 0.00)


| Location | 22,435,915 – 22,436,016 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -13.17 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
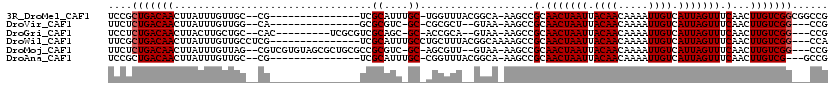
>3R_DroMel_CAF1 22435915 101 - 27905053 UCCGCUGACAACUUAUUUGUUGC--CG---------------UCGCAUUUGC-UGGUUUACGGCA-AAGCCGCAACUAAUUACAACAAAAUUGUCAUUAGUUUCAACUUGUCGGCGGCCG .((((((((((.......((((.--.(---------------(.((.(((((-((.....)))))-)))).))(((((((.((((.....)))).))))))).))))))))))))))... ( -35.81) >DroVir_CAF1 132509 95 - 1 UUCUCUGACAACUUAUUUGUUGG--CA---------------GCGCGUC-GC-CGCGCU--GUAA-AAGCCGCAACUAAUUACAACAAAAUUGUCAUUAGUUUCAACUUGUCGG---CCG ....(((((((.......(((.(--((---------------(((((..-..-))))))--))..-.))).(.(((((((.((((.....)))).))))))).)...)))))))---... ( -30.20) >DroGri_CAF1 110574 101 - 1 UCCUCUGACAACUUACUUGCUGC--CAC---------UCGCGUCGCAGC-GC-ACCGCA--GUAA-AAGCCGCAACUAAUUACAACAAAAUUGUCAUUAGUUUCAACUUGUCGG---CCG ....(((((((.....(((((((--...---------.(((......))-).-...)))--))))-.....(.(((((((.((((.....)))).))))))).)...)))))))---... ( -27.20) >DroWil_CAF1 73644 102 - 1 UUCGCUGACAACUUAUUUGUUGCCUCG---------------UCGCAUUUGCCUGCUUUACGGCAAAAGCCGCAACUAAUUACAACAAAAUUGUCAUUAGUUUCAACUUGUCGG---CCA ...((((((((.......((((....(---------------(.((.((((((........)))))).)).))(((((((.((((.....)))).))))))).)))))))))))---).. ( -30.11) >DroMoj_CAF1 161120 110 - 1 UUCUCUGACAACUUAUUUGUUAG--CGUCGUGUAGCGCUGCGCCGCGUC-GC-AGCGUU--GUAA-AAGCCGCAACUAAUUACAACAAAAUUGUCAUUAGUUUCAACUUGUCGG---CCG ....(((((((...........(--((.(.((((((((((((......)-))-))))))--))).-..).)))(((((((.((((.....)))).))))))).....)))))))---... ( -34.30) >DroAna_CAF1 75423 98 - 1 UCCGCUGACAACUUAUUUGUUGC--CG---------------UCGCAUUUGC-CGGUUUACGGCA-AAGCCGCAACUAAUUACAACAAAAUUGUCAUUAGUUUCAACUUGUCG---GCCG ...((((((((.......((((.--.(---------------(.((.(((((-((.....)))))-)))).))(((((((.((((.....)))).))))))).))))))))))---)).. ( -32.91) >consensus UCCGCUGACAACUUAUUUGUUGC__CG_______________UCGCAUC_GC_AGCGUU__GGAA_AAGCCGCAACUAAUUACAACAAAAUUGUCAUUAGUUUCAACUUGUCGG___CCG ....(((((((.................................((....))...................(.(((((((.((((.....)))).))))))).)...)))))))...... (-13.17 = -13.33 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:01 2006