
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,432,179 – 22,432,304 |
| Length | 125 |
| Max. P | 0.820321 |

| Location | 22,432,179 – 22,432,276 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -25.63 |
| Energy contribution | -25.22 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
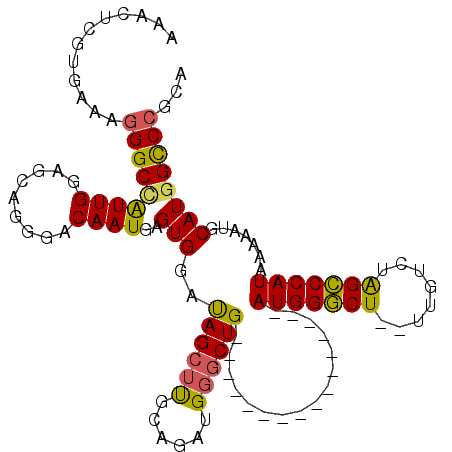
>3R_DroMel_CAF1 22432179 97 + 27905053 AAACUCGUCAAAGGGCCAUUGGAGCUGGGACAAUGAGUGGAUAGCUUGCAGAUGGGCUG----------------AUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCA ............(((((((..((((((...((.....))..))))))(((.((((((((----------------(..(...--)..)).))))))).....))))))))))... ( -36.50) >DroSec_CAF1 65712 97 + 1 AAACUCGUGAAAGGGCCAUUGGAGCUGGGACAAUGAGUGGAUAGCUUGCUGAUGGGCUG----------------AUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCA ............(((((((..((((((...((.....))..))))))((..((((((((----------------(..(...--)..)).)))))))......)))))))))... ( -34.40) >DroSim_CAF1 62103 97 + 1 AAACUCGUGAAAGGGCCAUUGGAGCUGGGACAAUGAGUGGAUAGCUUGCUGAUGGGCUG----------------AUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCA ............(((((((..((((((...((.....))..))))))((..((((((((----------------(..(...--)..)).)))))))......)))))))))... ( -34.40) >DroEre_CAF1 64122 113 + 1 AAACUCGGGAAAGGGCUAUUGGAGCAGGGACAAUGAGUGGAUAGCUCGCAGAUGGGCUGAUGGCUUGAUGGCCUGAUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCA .....((((...((((((((.(..((.......))..).))))))))(((.((((((((..(((.....((((.....))))--..))))))))))).....)))....)))).. ( -42.40) >DroYak_CAF1 63887 105 + 1 AAACUCGUGAAAGGGCCGUUGGAGCAGGGACAAUGAGUGGAUAGCUUGCAGAUGGGCUGAU--------GGCCUGAUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCA ............(((((((..((((..(..((.....))..).))))(((.((((((((((--------((((.....))))--..))).))))))).....))))))))))... ( -38.80) >DroAna_CAF1 71379 97 + 1 AAUCCCAUGAAAAGGCCAUUGGUGAACAGACAAUGAGUGGCCAG--UGCGGAUGGGCUG----------------AUGGCCUGAUGGUCUGGGCCAUAAAAAUGCAUAGUCCGCA .............(((((((.((.........)).)))))))..--(((((((..((..----------------(((((((........)))))))......))...))))))) ( -37.30) >consensus AAACUCGUGAAAGGGCCAUUGGAGCAGGGACAAUGAGUGGAUAGCUUGCAGAUGGGCUG________________AUGGGCU__UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCA ............(((((((((.........))))..(((..((((((......))))))................(((((((........))))))).......))))))))... (-25.63 = -25.22 + -0.41)


| Location | 22,432,179 – 22,432,276 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.37 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22432179 97 - 27905053 UGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAU----------------CAGCCCAUCUGCAAGCUAUCCACUCAUUGUCCCAGCUCCAAUGGCCCUUUGACGAGUUU ...((((((((((.....(((((((.((...--......)----------------)))))))).))).((((....((.....))...))))...)))))))............ ( -27.40) >DroSec_CAF1 65712 97 - 1 UGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAU----------------CAGCCCAUCAGCAAGCUAUCCACUCAUUGUCCCAGCUCCAAUGGCCCUUUCACGAGUUU ...(((((((........(((((((......--)))))))----------------..((......)).((((....((.....))...))))...)))))))............ ( -26.60) >DroSim_CAF1 62103 97 - 1 UGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAU----------------CAGCCCAUCAGCAAGCUAUCCACUCAUUGUCCCAGCUCCAAUGGCCCUUUCACGAGUUU ...(((((((........(((((((......--)))))))----------------..((......)).((((....((.....))...))))...)))))))............ ( -26.60) >DroEre_CAF1 64122 113 - 1 UGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAUCAGGCCAUCAAGCCAUCAGCCCAUCUGCGAGCUAUCCACUCAUUGUCCCUGCUCCAAUAGCCCUUUCCCGAGUUU .((((((...........(((((((......--)))))))..(((......)))....))))...((.((((.....((.....))....))))))...)).............. ( -29.90) >DroYak_CAF1 63887 105 - 1 UGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAUCAGGCC--------AUCAGCCCAUCUGCAAGCUAUCCACUCAUUGUCCCUGCUCCAACGGCCCUUUCACGAGUUU ...(((((.((((.....(((((((.((...--.(((.....))).--------.))))))))).))))(((.....((.....))....))).....)))))............ ( -30.50) >DroAna_CAF1 71379 97 - 1 UGCGGACUAUGCAUUUUUAUGGCCCAGACCAUCAGGCCAU----------------CAGCCCAUCCGCA--CUGGCCACUCAUUGUCUGUUCACCAAUGGCCUUUUCAUGGGAUU ((((((....((......((((((..........))))))----------------..))...))))))--..(((((.....((......))....)))))............. ( -23.50) >consensus UGCGGGCCAUGCAUUUUUAUGGGCUAGACAA__AGCCCAU________________CAGCCCAUCUGCAAGCUAUCCACUCAUUGUCCCAGCUCCAAUGGCCCUUUCACGAGUUU ...(((((((........(((((((........)))))))..................((......))............................)))))))............ (-20.23 = -20.37 + 0.14)



| Location | 22,432,214 – 22,432,304 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -24.36 |
| Energy contribution | -23.89 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22432214 90 + 27905053 AGUGGAUAGCUUGCAGAUGGGCUG----------------AUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCAUUGCAAUUUUGAUUGCGCUGCCCC-----C--GUU ...((...(((((((.((((((((----------------(..(...--)..)).))))))).....)))).)))..(((.((((((....)))))).)))..)-----)--... ( -30.50) >DroSec_CAF1 65747 90 + 1 AGUGGAUAGCUUGCUGAUGGGCUG----------------AUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCAUUGCAAUUUUGAUUGCGCUGCCCC-----C--GUU .(.((.((((....((.((((((.----------------(((((((--......)))))))..........))))))))..(((((....))))))))).)).-----)--... ( -30.40) >DroSim_CAF1 62138 90 + 1 AGUGGAUAGCUUGCUGAUGGGCUG----------------AUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCAUUGCAAUUUUGAUUGCGCUGCCCC-----C--GUU .(.((.((((....((.((((((.----------------(((((((--......)))))))..........))))))))..(((((....))))))))).)).-----)--... ( -30.40) >DroEre_CAF1 64157 106 + 1 AGUGGAUAGCUCGCAGAUGGGCUGAUGGCUUGAUGGCCUGAUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCAUUGCAAUUUUGAUUGCGAUGCCCC-----C--GUU .((((.....))))....(((((...((((....))))..(((((((--......)))))))..........)))))((((((((((....))))))))))...-----.--... ( -41.10) >DroYak_CAF1 63922 100 + 1 AGUGGAUAGCUUGCAGAUGGGCUGAU--------GGCCUGAUGGGCU--UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCAUUGCAAUUUUGAUUGCGCUGCCCC-----CCGGUU ..(((...(((((((.((((((((((--------((((.....))))--..))).))))))).....)))).)))..(((.((((((....)))))).)))...-----)))... ( -37.00) >DroAna_CAF1 71414 95 + 1 AGUGGCCAG--UGCGGAUGGGCUG----------------AUGGCCUGAUGGUCUGGGCCAUAAAAAUGCAUAGUCCGCAUUGCAAUUUUGAUUGGACCACCUCUUUCCC--AUU .((((.(((--(((((((..((..----------------(((((((........)))))))......))...))))))))))((((....))))..)))).........--... ( -35.00) >consensus AGUGGAUAGCUUGCAGAUGGGCUG________________AUGGGCU__UUGUCUAGCCCAUAAAAAUGCAUGGCCCGCAUUGCAAUUUUGAUUGCGCUGCCCC_____C__GUU ...((.............((((((................(((((((........))))))).........))))))(((.((((((....)))))).)))))............ (-24.36 = -23.89 + -0.47)



| Location | 22,432,214 – 22,432,304 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -23.51 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22432214 90 - 27905053 AAC--G-----GGGGCAGCGCAAUCAAAAUUGCAAUGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAU----------------CAGCCCAUCUGCAAGCUAUCCACU ...--.-----.((..((((((((....)))))..(((((((..((.......(((((((......--)))))))----------------))))))....))).)))..))... ( -29.11) >DroSec_CAF1 65747 90 - 1 AAC--G-----GGGGCAGCGCAAUCAAAAUUGCAAUGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAU----------------CAGCCCAUCAGCAAGCUAUCCACU ...--.-----.((..((((((((....)))))..(((((((..((.......(((((((......--)))))))----------------))))))....))).)))..))... ( -29.11) >DroSim_CAF1 62138 90 - 1 AAC--G-----GGGGCAGCGCAAUCAAAAUUGCAAUGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAU----------------CAGCCCAUCAGCAAGCUAUCCACU ...--.-----.((..((((((((....)))))..(((((((..((.......(((((((......--)))))))----------------))))))....))).)))..))... ( -29.11) >DroEre_CAF1 64157 106 - 1 AAC--G-----GGGGCAUCGCAAUCAAAAUUGCAAUGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAUCAGGCCAUCAAGCCAUCAGCCCAUCUGCGAGCUAUCCACU ..(--(-----.((((((.(((((....))))).))))((((...........(((((((......--)))))))..(((......)))....))))..)).))........... ( -36.00) >DroYak_CAF1 63922 100 - 1 AACCGG-----GGGGCAGCGCAAUCAAAAUUGCAAUGCGGGCCAUGCAUUUUUAUGGGCUAGACAA--AGCCCAUCAGGCC--------AUCAGCCCAUCUGCAAGCUAUCCACU ...(((-----(((((.(((((((....)))))...)).((((.((.......(((((((......--)))))))))))))--------....)))).))))............. ( -32.91) >DroAna_CAF1 71414 95 - 1 AAU--GGGAAAGAGGUGGUCCAAUCAAAAUUGCAAUGCGGACUAUGCAUUUUUAUGGCCCAGACCAUCAGGCCAU----------------CAGCCCAUCCGCA--CUGGCCACU ...--........(((((((((((....))))...((((((....((......((((((..........))))))----------------..))...))))))--..))))))) ( -25.70) >consensus AAC__G_____GGGGCAGCGCAAUCAAAAUUGCAAUGCGGGCCAUGCAUUUUUAUGGGCUAGACAA__AGCCCAU________________CAGCCCAUCUGCAAGCUAUCCACU ............((((...(((((....)))))((((((.....))))))...(((((((........)))))))..................)))).................. (-23.51 = -23.60 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:58 2006