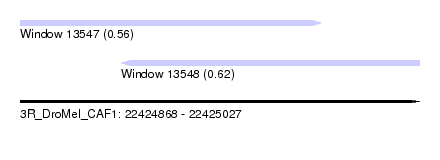
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,424,868 – 22,425,027 |
| Length | 159 |
| Max. P | 0.622098 |

| Location | 22,424,868 – 22,424,988 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -46.33 |
| Consensus MFE | -36.17 |
| Energy contribution | -35.65 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
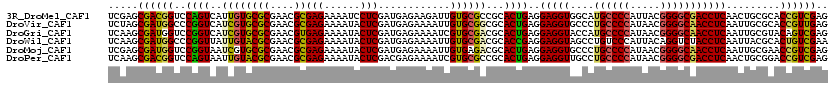
>3R_DroMel_CAF1 22424868 120 + 27905053 UCGAGCGACGGUCCAGUCAUUGUGCGCGAACGCGAGAAAAUCCUCGAUGAGAAGAUUGUGCGCCGCACUGAGGAGGUGGCAUGCCCCAUUACGGGGCGACCUCAACUGCGCACCGUCGAG .....((((((((((((....(((((.(..((((....((((.((.....)).)))).))))))))))....(((((....((((((.....))))))))))).)))).).))))))).. ( -48.70) >DroVir_CAF1 120001 120 + 1 UCUAGCGAUGGCCCGGUCAUCGUGCGCGAACGCGAGAAAAUACUCGAUGAGAAAAUUGUGCGGCGCACUGAGGAGGUGCCCUGCCCCAUAACGGGGCAACCUCAAUUGCGCACCGUUGAG ..((((((((((...))))))((((((((...((((......)))).((((..........((.(((((.....)))))))((((((.....))))))..)))).)))))))).)))).. ( -50.00) >DroGri_CAF1 98404 120 + 1 UCAAGCGAUGGUCCGGUCAUCGUGCGCGAACGUGAGAAAAUACUCGAUGAGAAAAUCGUGCGACGCACUGAGGAGGUACCAUGCCCCAUAACGGGGCAACCUCAAUUGCGUACAGUCGAG ((....(((((.....)))))((((((((.(....)......((((((......)))((((...)))).)))(((((....((((((.....)))))))))))..))))))))....)). ( -43.80) >DroWil_CAF1 61648 120 + 1 UCAAGCGAUGGCCCGGUUAUUGUACGCGAACGCGAGAAAAUACUCGAUGAGAAAAUUGUGCGACGCACCGAGGAGGUAGCCUGUCCCAUUACAGGUCUACCUCAAUUACGCACUGUCGAA .....((((((..((((..((((((((....))(((......)))............))))))...))))..((((((((((((......))))).))))))).........)))))).. ( -39.70) >DroMoj_CAF1 147807 120 + 1 UCGAGCGAUGGUCCGGUAAUCGUGCGCGAACGCGAGAAAAUACUCGAUGAGAAAAUUGUGAGACGCACUGAGGAGGUGCCCUGCCCCAUAACGGGGCAACCUCAAUUGCGAACCGUCGAG .....(((((((.((.((((.((((((..(((((((......))))..........)))..).)))))(((((...(((((((........))))))).))))))))))).))))))).. ( -43.80) >DroPer_CAF1 78715 120 + 1 UCAAGCGACGGUCCAGUAAUUGUACGCGAACGCGAGAAAAUACUCGACGAGAAAAUCGUGCGCCGCACUGAGGAGGUUGCCUGCCCCAUAACGGGGCGACCUCAACUGCGGACCGUCGAG .....(((((((((.(((.......(((..((((((......))).((((.....))))))).)))..((((((((...)))(((((.....)))))..)))))..)))))))))))).. ( -52.00) >consensus UCAAGCGAUGGUCCGGUCAUCGUGCGCGAACGCGAGAAAAUACUCGAUGAGAAAAUUGUGCGACGCACUGAGGAGGUGCCCUGCCCCAUAACGGGGCAACCUCAAUUGCGCACCGUCGAG .....((((((..((((..((((((((....))(((......)))............))))))...))))..(((((....((((((.....))))))))))).........)))))).. (-36.17 = -35.65 + -0.52)

| Location | 22,424,908 – 22,425,027 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.23 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22424908 119 - 27905053 G-CUGGCUCUAACCUCCGUCUCGAUGAUUUUCUCAAUGUACUCGACGGUGCGCAGUUGAGGUCGCCCCGUAAUGGGGCAUGCCACCUCCUCAGUGCGGCGCACAAUCUUCUCAUCGAGGA (-..((......))..).(((((((((.........(((.....)))((((((.(((((((..(((((.....))))).........)))))..)).)))))).......))))))))). ( -39.40) >DroVir_CAF1 120041 114 - 1 ------AGCUAACCUCCGUCUCGAUGAUUUUCUCAAUGUACUCAACGGUGCGCAAUUGAGGUUGCCCCGUUAUGGGGCAGGGCACCUCCUCAGUGCGCCGCACAAUUUUCUCAUCGAGUA ------.............((((((((.........((....)).((((((((....(((((((((((.....))))))....)))))....))))))))..........)))))))).. ( -41.60) >DroGri_CAF1 98444 114 - 1 ------AGCUAACCUCCGUCUCGAUGAUUUUCUCAAUGUACUCGACUGUACGCAAUUGAGGUUGCCCCGUUAUGGGGCAUGGUACCUCCUCAGUGCGUCGCACGAUUUUCUCAUCGAGUA ------.............((((((((.....((...((((......))))(((.((((((.((((((.....))))))........))))))))).......)).....)))))))).. ( -37.30) >DroWil_CAF1 61688 117 - 1 ---UAGUUCUAACCUCUGUUUCGAUGAUUUUCUCAAUGUAUUCGACAGUGCGUAAUUGAGGUAGACCUGUAAUGGGACAGGCUACCUCCUCGGUGCGUCGCACAAUUUUCUCAUCGAGUA ---................((((((((.........(((.....)))(((((((...(((((((.(((((......)))))))))))).....)))).))).........)))))))).. ( -35.50) >DroMoj_CAF1 147847 114 - 1 ------CGCUAACCUCCGUCUCGAUGAUUUUCUCAAUGUACUCGACGGUUCGCAAUUGAGGUUGCCCCGUUAUGGGGCAGGGCACCUCCUCAGUGCGUCUCACAAUUUUCUCAUCGAGUA ------.............((((((((..........((.....))((..((((.((((((.(((((.(((....))).)))))...))))))))))..)).........)))))))).. ( -35.80) >DroPer_CAF1 78755 120 - 1 GUAUGUAGCUAACCUCCGUUUCGAUGAUUUUCUCAAUGUACUCGACGGUCCGCAGUUGAGGUCGCCCCGUUAUGGGGCAGGCAACCUCCUCAGUGCGGCGCACGAUUUUCUCGUCGAGUA ................((((..((......))..))))((((((((((((((((.((((((..(((((.....))))).(....)..))))))))))).))..(.....).))))))))) ( -40.90) >consensus ______AGCUAACCUCCGUCUCGAUGAUUUUCUCAAUGUACUCGACGGUGCGCAAUUGAGGUUGCCCCGUUAUGGGGCAGGCCACCUCCUCAGUGCGUCGCACAAUUUUCUCAUCGAGUA ...................((((((((..........((((......))))(((.((((((..(((((.....)))))(((...))))))))))))..............)))))))).. (-31.40 = -31.23 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:54 2006