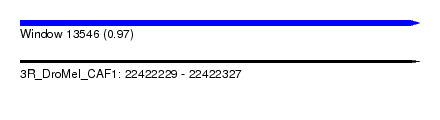
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,422,229 – 22,422,327 |
| Length | 98 |
| Max. P | 0.968540 |

| Location | 22,422,229 – 22,422,327 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22422229 98 + 27905053 ---GCUUGGAUUAUUGU-CGGAGGCGAGCGGUCGAGUGCCGGGGCAAAGUUCAUUUAAAAGUCGAGAGGACAAAUGAAUGAAUAAAAGGAGCAAGGAGGCAA---------- ---((((......((((-(...((((..(....)..))))..))))).(((((((((...(((.....)))...))))))))).....))))..........---------- ( -25.40) >DroVir_CAF1 116908 102 + 1 -----UGCCAGCAUGUC-UGGUGGUGCCCUCUGGAGUGCCUGCGCAAAGUUCAUUUAAAAGUCGAAAGGACAAAUGAAUGAAUAAAGCGAGCCGAGCAAAUGGCAGCG---- -----(((((...((..-((((((..(((...)).)..))..(((...(((((((((...(((.....)))...)))))))))...))).))))..))..)))))...---- ( -34.90) >DroSec_CAF1 55759 98 + 1 ---GCUUGGAUUAUUGU-UGGAGGCGCGCGGUCGAGUGCCGGGGCAAAGUUCAUUUAAAAGUCGAGAGGACAAAUGAAUGAAUAAAAGGAGCAAGGAGGCAA---------- ---((((......((((-(...(((((........)))))..))))).(((((((((...(((.....)))...))))))))).....))))..........---------- ( -25.90) >DroSim_CAF1 52591 98 + 1 ---GCUCGGAUUAUUGU-UGGAGGCGCGCGGUCGAGUGCCGGGGCAAAGUUCAUUUAAAAGUCGAGAGGACAAAUGAAUGAAUAAAAGGAGCAAGGAGGCAA---------- ---((((......((((-(...(((((........)))))..))))).(((((((((...(((.....)))...))))))))).....))))..........---------- ( -28.10) >DroEre_CAF1 54092 98 + 1 ---GCUUGGAUUAUUGU-UGGAGGCGCUCGGUCGAGUGCAGGGGCAAAGUUCAUUUAAAAGUCGAGAGGACAAAUGAAUGAAUAAAGGGAGCAAGGAGGCAA---------- ---((((......((((-(....((((((....))))))...))))).(((((((((...(((.....)))...))))))))).....))))..........---------- ( -26.50) >DroPer_CAF1 75826 108 + 1 CGGGC---GACCAUUGUAUGUCGGUACGCGCUCGAGUGUCAGAGCAAAGUUCAUUUAAAAGUCGAGGGGACAAAUGAA-GAAUAAGAGGAGCGGAGCGCCAAGCAAAUGCCA ..(((---(....((((.((.((.(.(((((((........))))....(((((((....(((.....))))))))))-...........))).).)).)).)))).)))). ( -27.80) >consensus ___GCUUGGAUUAUUGU_UGGAGGCGCGCGGUCGAGUGCCGGGGCAAAGUUCAUUUAAAAGUCGAGAGGACAAAUGAAUGAAUAAAAGGAGCAAGGAGGCAA__________ ...((((......((((.....(((((.(....).)))))...)))).(((((((((...(((.....)))...))))))))).....)))).................... (-17.04 = -17.98 + 0.95)

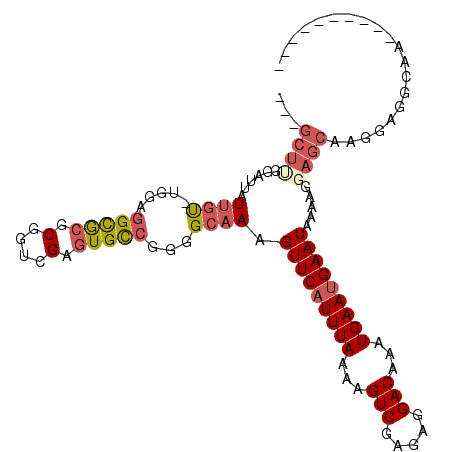
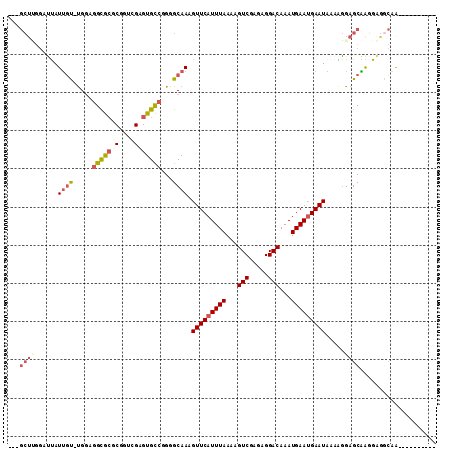
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:52 2006