| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,406,084 – 22,406,224 |
| Length | 140 |
| Max. P | 0.971445 |

| Location | 22,406,084 – 22,406,184 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 65.40 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22406084 100 - 27905053 UUAUAUUGGAGGUGGCAGCAGGUGGUGUGGGCGACGAGGCGCCAUGCACCAUGGCAGCAGCAGCUGCCAGGACAGAUGGGCCAGUUGGCUGU--UGU----GCAUC ............(((((((..((((((((((((......)))).)))))))).((....)).)))))))(.((((...((((....)))).)--)))----.)... ( -42.20) >DroPse_CAF1 57566 79 - 1 U------------------GGUGGGUACGGGCGAUGGGGCGCCAUGCACCAGGGCAGCAGCAGCUGCCAGACCCGUCUGACUCGUUGGCUCC---------GCCUC .------------------(((((((((((((((((((((.....)).....((((((....))))))...)))))).).)))))..)).))---------))).. ( -37.80) >DroEre_CAF1 37884 100 - 1 UUGUAUUGGAGGUGGCAGCAGGAGGUGUGGGCGACGCAGCGCCGUGCACCAUGGCAGCAGCGGCUGCCAGAACCGAUUGGCCAGUUGGCUGC--UGU----GUGUC ..............(((((((..((((((((((......)))).)))))).(((((((....)))))))...((((((....))))))))))--)))----..... ( -43.70) >DroWil_CAF1 40631 100 - 1 U--GAUUGGUGUCGGU----GUGGGUGUGGGAGAUGGUGUUCCAUGCACCAAGGCAGCAGCAACAGCCAAGCCGGAUUGGCUAGUUGGUUCUGGUGUGGUAGCAUC (--(.(((.((((...----.(((((((((((.......)))))))).))).)))).)))))...((((.(((((((..(....)..).)))))).))))...... ( -34.90) >DroAna_CAF1 43047 82 - 1 U---ACUCGUG------------GGCGUCGGUGAGGCGGCUCCGUGCACCAGGGCAGCAGCAGCUGCCAGGCCAGUUUGGCUCGUUGCC-----UGU----GCAUC .---...((..------------((((.(((((..(((....))).))))..((((((....)))))).((((.....)))).).))))-----..)----).... ( -36.00) >DroPer_CAF1 58364 79 - 1 U------------------GGUGGGUACGGGCGAUGGGGCGCCAUGCACCAGGGCAGCAGCAGCUGCCAGACCCGUCUGACUCGUUGGCUCC---------GCCUC .------------------(((((((((((((((((((((.....)).....((((((....))))))...)))))).).)))))..)).))---------))).. ( -37.80) >consensus U___AUUGG_G_________GUGGGUGUGGGCGAUGGGGCGCCAUGCACCAGGGCAGCAGCAGCUGCCAGACCCGAUUGGCUAGUUGGCUCC__UGU____GCAUC .......................((((((((((......)))).))))))..((((((....))))))...................................... (-21.07 = -21.65 + 0.58)

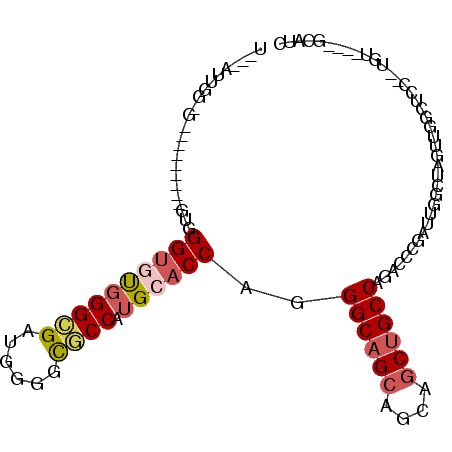

| Location | 22,406,113 – 22,406,224 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.10 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22406113 111 + 27905053 CCUGGCAGCUGCUGCUGCCAUGGUGCAUGGCGCCUCGUCGCCCACACCACCUGCUGCCACCUCCAAUAUAACUCCCUUACCCACAGCCGCGGCAGUAGCAUCUGCUGAUUU ...(((((.((((((((((.(((((...((((......))))..)))))...((((...........................))))...)))))))))).)))))..... ( -42.23) >DroPse_CAF1 57592 84 + 1 UCUGGCAGCUGCUGCUGCCCUGGUGCAUGGCGCCCCAUCGCCCGUACCCACC------------------AC------ACCUAC-GC--CGCCUGUGGCAGCCGCUGAGUU .....((((.(((((..(...((((...((((......))))((((......------------------..------...)))-).--)))).)..))))).)))).... ( -30.40) >DroSec_CAF1 39764 111 + 1 CCUGGCAGCUGCUGCUGCCAUGGUGCAUGGCGCCUCGUCGCCCACACCACCUGCUGCCACCUCCAAUACAACUCCCUUACCCACAGCCGCGCCAGUGGCAUCUGCUGAUUU ..(((((((.((....))..(((((...((((......))))..)))))...)))))))..........................(((((....)))))............ ( -34.70) >DroEre_CAF1 37913 111 + 1 UCUGGCAGCCGCUGCUGCCAUGGUGCACGGCGCUGCGUCGCCCACACCUCCUGCUGCCACCUCCAAUACAACUCCAUUACCCACAGCCGCGCCAGUGGCAUCUGCUGAUUU ..(((((((.((....))...((((...((((......))))..))))....)))))))..........................(((((....)))))............ ( -33.40) >DroAna_CAF1 43073 96 + 1 CCUGGCAGCUGCUGCUGCCCUGGUGCACGGAGCCGCCUCACCGACGCC------------CACGAGU---ACACCCAUUCCGACCCCCGUGCCAGUGGCAUCCGCGGACUU ...((((((....))))))..(((.(.(((((((((..(((.......------------...((((---......))))........)))...))))).)))).).))). ( -31.17) >DroPer_CAF1 58390 84 + 1 UCUGGCAGCUGCUGCUGCCCUGGUGCAUGGCGCCCCAUCGCCCGUACCCACC------------------AC------ACCUAC-GC--CGCCUGCGGCAGCCGCUGACUU ...((((((.((((((((...((((...((((......))))((((......------------------..------...)))-).--)))).)))))))).)))).)). ( -33.10) >consensus CCUGGCAGCUGCUGCUGCCAUGGUGCAUGGCGCCCCGUCGCCCACACCACCU________CUCCAAU___ACUCCCUUACCCACAGCCGCGCCAGUGGCAUCCGCUGAUUU ...((((((....))))))..((((...((((......))))..))))...................................((((...((.....))....)))).... (-24.38 = -24.68 + 0.31)

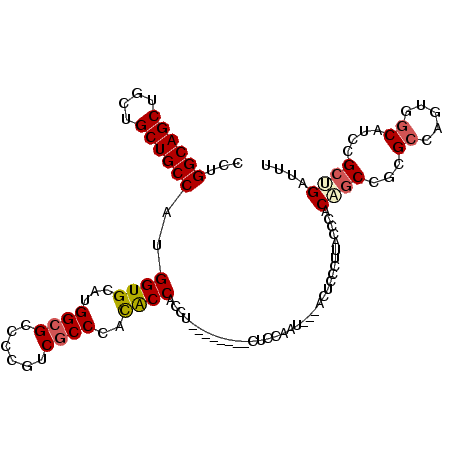

| Location | 22,406,113 – 22,406,224 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.10 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -28.22 |
| Energy contribution | -29.05 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22406113 111 - 27905053 AAAUCAGCAGAUGCUACUGCCGCGGCUGUGGGUAAGGGAGUUAUAUUGGAGGUGGCAGCAGGUGGUGUGGGCGACGAGGCGCCAUGCACCAUGGCAGCAGCAGCUGCCAGG ......((((.((((.(((((((.(((.(.((((.........)))).).))).)).....((((((((((((......)))).))))))))))))).)))).)))).... ( -46.00) >DroPse_CAF1 57592 84 - 1 AACUCAGCGGCUGCCACAGGCG--GC-GUAGGU------GU------------------GGUGGGUACGGGCGAUGGGGCGCCAUGCACCAGGGCAGCAGCAGCUGCCAGA .(((..(((.(((((...))))--))-)).)))------.(------------------((((.((...((((......)))))).))))).((((((....))))))... ( -36.00) >DroSec_CAF1 39764 111 - 1 AAAUCAGCAGAUGCCACUGGCGCGGCUGUGGGUAAGGGAGUUGUAUUGGAGGUGGCAGCAGGUGGUGUGGGCGACGAGGCGCCAUGCACCAUGGCAGCAGCAGCUGCCAGG ......((...(((((((...((((((.(.......).))))))......)))))))))..((((((((((((......)))).))))))))((((((....))))))... ( -45.50) >DroEre_CAF1 37913 111 - 1 AAAUCAGCAGAUGCCACUGGCGCGGCUGUGGGUAAUGGAGUUGUAUUGGAGGUGGCAGCAGGAGGUGUGGGCGACGCAGCGCCGUGCACCAUGGCAGCAGCGGCUGCCAGA ......((...(((((((...((((((.((.....)).))))))......)))))))))....((((((((((......)))).)))))).(((((((....))))))).. ( -44.80) >DroAna_CAF1 43073 96 - 1 AAGUCCGCGGAUGCCACUGGCACGGGGGUCGGAAUGGGUGU---ACUCGUG------------GGCGUCGGUGAGGCGGCUCCGUGCACCAGGGCAGCAGCAGCUGCCAGG ..((.((((((.(((.((..(((.((.(((...(((((...---.))))).------------))).)).))).)).))))))))).))...((((((....))))))... ( -44.90) >DroPer_CAF1 58390 84 - 1 AAGUCAGCGGCUGCCGCAGGCG--GC-GUAGGU------GU------------------GGUGGGUACGGGCGAUGGGGCGCCAUGCACCAGGGCAGCAGCAGCUGCCAGA ......((((...)))).((((--((-((.(.(------((------------------..((((((..((((......)))).))).)))..))).).)).))))))... ( -36.00) >consensus AAAUCAGCAGAUGCCACUGGCGCGGCUGUGGGUAAGGGAGU___AUUGGAG________AGGGGGUGUGGGCGACGAGGCGCCAUGCACCAGGGCAGCAGCAGCUGCCAGA ....((((...((((...))))..))))...................................((((((((((......)))).))))))..((((((....))))))... (-28.22 = -29.05 + 0.83)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:49 2006