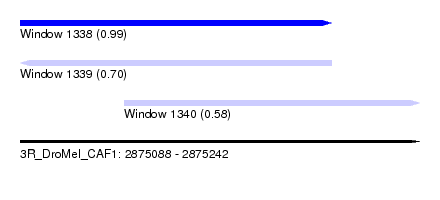
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,875,088 – 2,875,242 |
| Length | 154 |
| Max. P | 0.993617 |

| Location | 2,875,088 – 2,875,208 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -28.62 |
| Energy contribution | -29.25 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
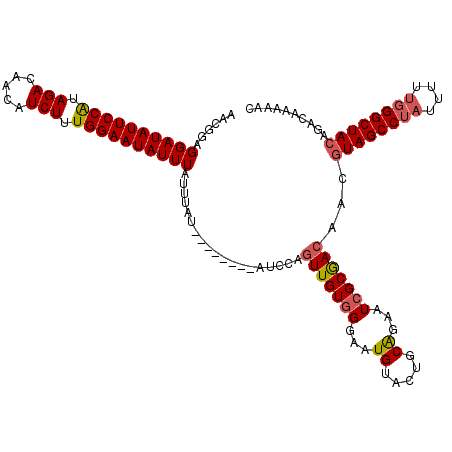
>3R_DroMel_CAF1 2875088 120 + 27905053 AACGGAGGAUAUUCCAUAGACAACAUCUUUGGAAUAUUUAUUUAUGUAUGUAUAUCCAGUUGUGGGAAUGUACUGCAGAAUCGCGACAACGUAGCCUAUUUUUGGGCUACCCACAAAAAC ...(((((((((((((.(((.....))).))))))))))...((((....)))))))..(((((((..(((...((......)).)))...(((((((....)))))))))))))).... ( -36.60) >DroSec_CAF1 6212 113 + 1 AACGGAGGAUAUUCCAUAGACAACAUCUUUGGAAUAUUUAUUUAUA-------AUCCAGUUGUGGGAAUGUACUGCGGAAUCGCGACAACGUAGCCUAUUUUCGGGCUACAGACAAAAAC ...(((((((((((((.(((.....))).)))))))))).......-------.))).(((((((...((.....))...)))))))...(((((((......))))))).......... ( -33.80) >DroSim_CAF1 8440 111 + 1 AACGGAGGAUAUUCCAUAGACAACAUCUUUGGAAUAUUUAUUUAU---------UCCAGUUGUGGGAAUGUACUGCGGAAUCGCGACAACGUAGCCUAUUUUUGGGCUACAGACAAAAAC ..((.((.(((((((....(((((.....(((((((......)))---------))))))))).))))))).)).)).............((((((((....)))))))).......... ( -36.00) >DroEre_CAF1 7183 112 + 1 AACGGAGGAUAUUCCGAAGACAACAUCUUUGGAAUAUUUAUUGAU--------CUCGAGUUGUGGGAUUGUACAGCAGAAUAGCAAAAACAUAGCCUAUUUUUCGGCUACCGACGAAAAU ..(((.((((((((((((((.....))))))))))))))......--------.(((((..(((((..(((...((......))....)))...)))))..)))))...)))........ ( -32.50) >consensus AACGGAGGAUAUUCCAUAGACAACAUCUUUGGAAUAUUUAUUUAU________AUCCAGUUGUGGGAAUGUACUGCAGAAUCGCGACAACGUAGCCUAUUUUUGGGCUACAGACAAAAAC ......((((((((((.(((.....))).))))))))))...................(((((((...((.....))...)))))))...((((((((....)))))))).......... (-28.62 = -29.25 + 0.62)

| Location | 2,875,088 – 2,875,208 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2875088 120 - 27905053 GUUUUUGUGGGUAGCCCAAAAAUAGGCUACGUUGUCGCGAUUCUGCAGUACAUUCCCACAACUGGAUAUACAUACAUAAAUAAAUAUUCCAAAGAUGUUGUCUAUGGAAUAUCCUCCGUU ....((((((((((((........))))))......(((....))).........))))))..((((((......))).....((((((((.((((...)))).))))))))..)))... ( -29.90) >DroSec_CAF1 6212 113 - 1 GUUUUUGUCUGUAGCCCGAAAAUAGGCUACGUUGUCGCGAUUCCGCAGUACAUUCCCACAACUGGAU-------UAUAAAUAAAUAUUCCAAAGAUGUUGUCUAUGGAAUAUCCUCCGUU ((((..((((((((((........))))))(((((.(((....))).(........)))))).))))-------...))))..((((((((.((((...)))).))))))))........ ( -27.50) >DroSim_CAF1 8440 111 - 1 GUUUUUGUCUGUAGCCCAAAAAUAGGCUACGUUGUCGCGAUUCCGCAGUACAUUCCCACAACUGGA---------AUAAAUAAAUAUUCCAAAGAUGUUGUCUAUGGAAUAUCCUCCGUU ......((((((((((........))))))......(((....))))).))(((((.......)))---------))......((((((((.((((...)))).))))))))........ ( -26.80) >DroEre_CAF1 7183 112 - 1 AUUUUCGUCGGUAGCCGAAAAAUAGGCUAUGUUUUUGCUAUUCUGCUGUACAAUCCCACAACUCGAG--------AUCAAUAAAUAUUCCAAAGAUGUUGUCUUCGGAAUAUCCUCCGUU ..(((((...((((((........))))))((....((......))...))............))))--------).......(((((((.(((((...))))).)))))))........ ( -23.20) >consensus GUUUUUGUCGGUAGCCCAAAAAUAGGCUACGUUGUCGCGAUUCCGCAGUACAUUCCCACAACUGGAU________AUAAAUAAAUAUUCCAAAGAUGUUGUCUAUGGAAUAUCCUCCGUU ..........((((((........))))))(((((.(((....))).(........)))))).....................((((((((.((((...)))).))))))))........ (-20.69 = -21.12 + 0.44)

| Location | 2,875,128 – 2,875,242 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
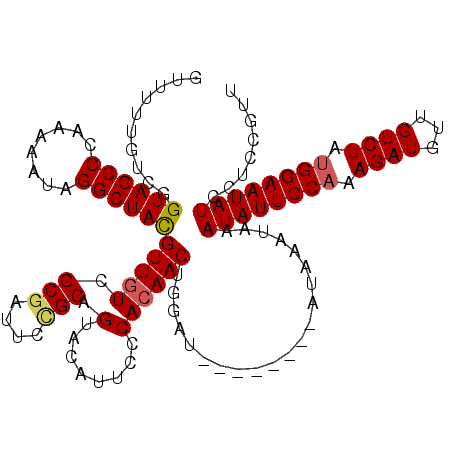
>3R_DroMel_CAF1 2875128 114 + 27905053 UUUAUGUAUGUAUAUCCAGUUGUGGGAAUGUACUGCAGAAUCGCGACAACGUAGCCUAUUUUUGGGCUACCCACAAAAACAACCUCUA----AGAUCACAUUCCUGUCUA--CAAAAGAU ..................((.(..(((((((..(((......))).....((((((((....))))))))..................----.....)))))))..)..)--)....... ( -25.60) >DroSec_CAF1 6252 109 + 1 UUUAUA-------AUCCAGUUGUGGGAAUGUACUGCGGAAUCGCGACAACGUAGCCUAUUUUCGGGCUACAGACAAAAACAACCUCUG----AGAUCACAUUCCUGUCUUUACAAAAGAU ......-------..........((((((((..((((....)))).....(((((((......)))))))..................----.....))))))))(((((.....))))) ( -26.10) >DroSim_CAF1 8480 109 + 1 UUUAU---------UCCAGUUGUGGGAAUGUACUGCGGAAUCGCGACAACGUAGCCUAUUUUUGGGCUACAGACAAAAACAACCUAUGUAUAAGAUCACAUUCCUGACUU--CAAAAGAU .....---------...((((..((((((((..((((....)))).....((((((((....))))))))...........................)))))))))))).--........ ( -27.80) >DroEre_CAF1 7223 106 + 1 UUGAU--------CUCGAGUUGUGGGAUUGUACAGCAGAAUAGCAAAAACAUAGCCUAUUUUUCGGCUACCGACGAAAAUAAGCCCAA----AGAUCACAUUCCUGUCUA--CAACAGAG .((((--------((.(((..(((((..(((...((......))....)))...)))))..)))((((.............))))...----)))))).....((((...--..)))).. ( -22.82) >consensus UUUAU________AUCCAGUUGUGGGAAUGUACUGCAGAAUCGCGACAACGUAGCCUAUUUUUGGGCUACAGACAAAAACAACCUCUA____AGAUCACAUUCCUGUCUA__CAAAAGAU .......................((((((((...((......))......((((((((....))))))))...........................))))))))............... (-20.60 = -21.60 + 1.00)

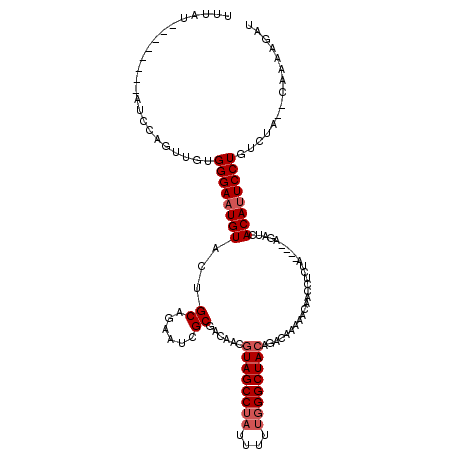
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:30 2006