
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,364,424 – 22,364,553 |
| Length | 129 |
| Max. P | 0.757380 |

| Location | 22,364,424 – 22,364,522 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -34.49 |
| Energy contribution | -35.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22364424 98 + 27905053 UCCCACCGAAUGGCCUGGUCCUAAGCUCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAGGGCCAUUCCCCCA .......((((((((....((..((((.....))))...))((((((..((....((((((....))))))...))...))))))))))))))..... ( -38.50) >DroSec_CAF1 5639 95 + 1 UCCCACCGAAUGGCCUGGUCCUAAGCUCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGGCCAGGGCCAUUCCC--- .......((((((((((..(((........)))..)..((((((((...((....((((((....))))))...)))))))))))))))))))..--- ( -41.70) >DroSim_CAF1 1096 95 + 1 UCCCACCGAAUGGCCUGGUCCUAAGCUCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAGGGCCAUUCCC--- .......((((((((....((..((((.....))))...))((((((..((....((((((....))))))...))...))))))))))))))..--- ( -38.50) >DroEre_CAF1 1183 98 + 1 UCCCACCGAAUGGCCUGGUCCUAAGCUCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAGGGCCAUUCCUUCA .......(((.((..(((((((..((....(((((..(((.(((((...((.....))...)))))...)))..)))))))..)))))))..))))). ( -38.60) >DroYak_CAF1 1190 98 + 1 UCCCACCGAAUGGCCUGGUCCUAAGCUCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAUGGCCAUUCCUCCA .......(((((((((((......((....(((((..(((.(((((...((.....))...)))))...)))..)))))))))).))))))))..... ( -37.30) >DroAna_CAF1 10814 97 + 1 UCCCGGCG-AUGGCCUGGCCCUAAGCUCAAGGGGUUGAUGGCUAGGCACGUAAUUUGCAUCCCCGGGUGCGAUGACCCCGCCCAGGGCCAGUUUUCCG .(((((((-(((((((((((...((((.....))))...)))))))).)))...(((((((....)))))))......))))..)))........... ( -40.10) >consensus UCCCACCGAAUGGCCUGGUCCUAAGCUCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAGGGCCAUUCCCCCA .......(((((((((((((...((((.....))))...))))((((..((....((((((....))))))...))...)))).)))))))))..... (-34.49 = -35.05 + 0.56)

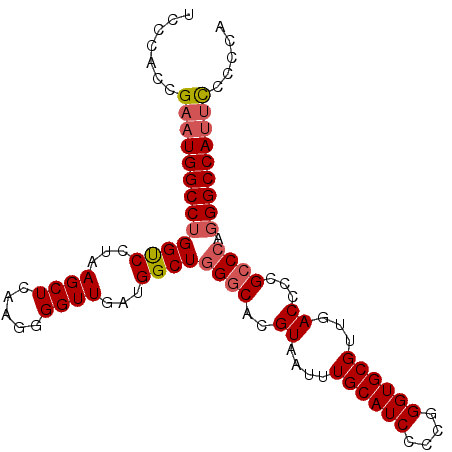

| Location | 22,364,450 – 22,364,553 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22364450 103 + 27905053 UCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAGGGCCAUUCCCCCAUCUCCUGCACCCCCUGUUCCUC-------CUUA--UGGCG .((.(((((.(((((((((((..((....((((((....))))))...))...))))))((......)))))))......))))).)).....(-------(...--.)).. ( -34.10) >DroSec_CAF1 5665 91 + 1 UCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGGCCAGGGCCAUUCCC--------------CCCUGUUCCUC-------CUUGUACAGCG ...(((((..(((((((.(((.((.....((((((....)))))).......)))))..)))))))..)--------------)))).......-------........... ( -35.60) >DroSim_CAF1 1122 91 + 1 UCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAGGGCCAUUCCC--------------CCCUAUUCCUC-------CUUGUACAGCG ...(((((..(((((((((((..((....((((((....))))))...))...))))..)))))))..)--------------)))).......-------........... ( -36.60) >DroYak_CAF1 1216 109 + 1 UCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAUGGCCAUUCCUCCACCUCG---ACAUCCAUCUCCUCCAUCUAUCUCCUCUAUCG ((.(((((..(((((((((((..((....((((((....))))))...))...))))..)))))))...)).))).)---)............................... ( -31.90) >DroAna_CAF1 10839 94 + 1 UCAAGGGGUUGAUGGCUAGGCACGUAAUUUGCAUCCCCGGGUGCGAUGACCCCGCCCAGGGCCAGUUUUCCGCCACA---GCCACAA--UCGCC-------CUCCU------ ....(((((((.(((((.(((..((...(((((((....)))))))..))(((.....)))..........)))..)---)))))))--)).))-------.....------ ( -33.80) >consensus UCAAGGGGUUGAUGGCUGGGCACGUAAUUUGCAUCCCCGGGUGCGUUGACCCCGCCCAGGGCCAUUCCCCC__C_C_____C_CCCUGUUCCUC_______CUUCU_CAGCG ....((((..(((((((((((..((....((((((....))))))...))...))))..))))))))))).......................................... (-25.04 = -25.48 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:33 2006