
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,362,693 – 22,362,883 |
| Length | 190 |
| Max. P | 0.922181 |

| Location | 22,362,693 – 22,362,803 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.91 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -23.10 |
| Energy contribution | -25.85 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22362693 110 + 27905053 CUUCGCUGU--------GGAGAGGAAGCAACAAAUGCGAUGCGUUGCGUUGC--GUUUCUUGUUUCUUUUAACUAACUACGUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAA ...((((.(--------((...((((((.....(((.((((((((((((..(--((.....(((......))).....)))..)))))))))))).))).)).))))..))))))).... ( -37.60) >DroSim_CAF1 74393 106 + 1 CUUCGCUGU--------GGAGAGGAAGCAACAAAUGCGAUGCGUUGCGUUGC--GUUUCUUGUUUCGUUUAA----CUACGUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAA ..((((.((--------(((((.(((((((.(((((((((((...)))))))--)))).))))))).)))..----))))))))..............(((((((.......))))))). ( -36.80) >DroEre_CAF1 73196 109 + 1 CUUCGUUGUCGAGUUGUCGAGAGGAAGCAACAAAUGCG-----UUGCGUUUC--GUUUCUUCUUCCUUUUAA----CUACGUGAUGCAAUGCGUUUAAUGGCAUUCCAACCAAGUGCCAA ((((.((.((((....)))).))))))....(((((((-----(((((((.(--((................----..))).))))))))))))))..(((((((.......))))))). ( -33.67) >DroYak_CAF1 72735 111 + 1 CUUCGUUGUCGAGUUGUCGAGAGGAAGCAACAAAUGCG-----UUGCGUUUCUUGUUCCUUUUUGUUUUUAA----CUACGUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAA ((((.((.((((....)))).))))))....(((((((-----(((((((...(((................----..))).))))))))))))))..(((((((.......))))))). ( -30.47) >consensus CUUCGCUGU________CGAGAGGAAGCAACAAAUGCG_____UUGCGUUGC__GUUUCUUGUUUCUUUUAA____CUACGUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAA ((((.((.(((......))).))))))....(((((((.(((((..(((.............................)))..))))).)))))))..(((((((.......))))))). (-23.10 = -25.85 + 2.75)

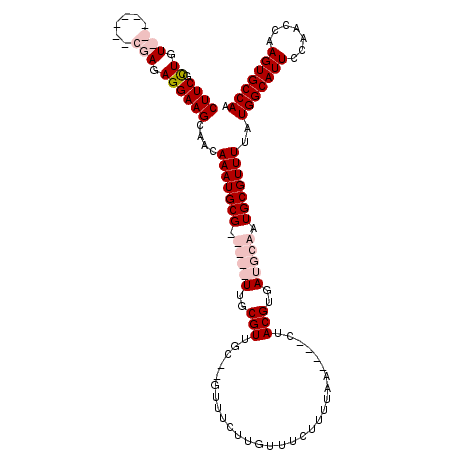
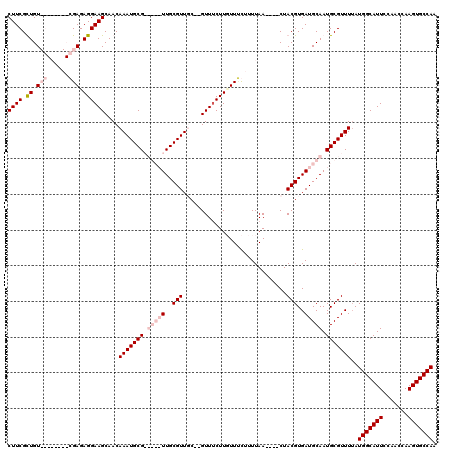
| Location | 22,362,763 – 22,362,883 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22362763 120 + 27905053 GUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAAUUAGAAUUGAUUAUUAUUCAUUAUUUCGCUUAUUACCCACACAGGCGCGUGUUUGUGCGUUUGUGUGUAAGCGGCUCUCU ....((.(((((........))))).))....((.(((.....(((.((((........)))).)))(((((.....((((((((((((......)))))))))))))))))))).)).. ( -35.10) >DroSim_CAF1 74459 120 + 1 GUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAAUUAGAAUUGAUUAUUAUUCAUUAUUUCGCUUAUUACCCACACAGGCGCGUGUUCGUGCGUUUGUGUGUAAGCGGCUCUCU ....((.(((((........))))).))....((.(((.....(((.((((........)))).)))(((((.....((((((((((((......)))))))))))))))))))).)).. ( -35.10) >DroEre_CAF1 73265 120 + 1 GUGAUGCAAUGCGUUUAAUGGCAUUCCAACCAAGUGCCAAUUAGAAUUGAUUAUUAUUCAUUAUUUCGCUUAUUACCCACACAGGCGCGUGUUUGUGCGUUUGUGUGUAAGCGGCUCUCU (((((.((((...((((((((((((.......)))))).)))))))))))))))...........(((((((.....((((((((((((......)))))))))))))))))))...... ( -35.30) >DroYak_CAF1 72806 120 + 1 GUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAAUUAGAAUUGAUUAUUAUUCAUUAUUUCGCUUAUUACCCACACAGGCGCGUGUUUGUGCGUUUGUGUGUAAGCGGCUCUCU ....((.(((((........))))).))....((.(((.....(((.((((........)))).)))(((((.....((((((((((((......)))))))))))))))))))).)).. ( -35.10) >consensus GUGAUGCAAUGCGUUUUAUGGCAUUCCAACCAAGUGCCAAUUAGAAUUGAUUAUUAUUCAUUAUUUCGCUUAUUACCCACACAGGCGCGUGUUUGUGCGUUUGUGUGUAAGCGGCUCUCU ....((.(((((........))))).))....((.(((.....(((.((((........)))).)))(((((.....((((((((((((......)))))))))))))))))))).)).. (-35.10 = -35.10 + 0.00)

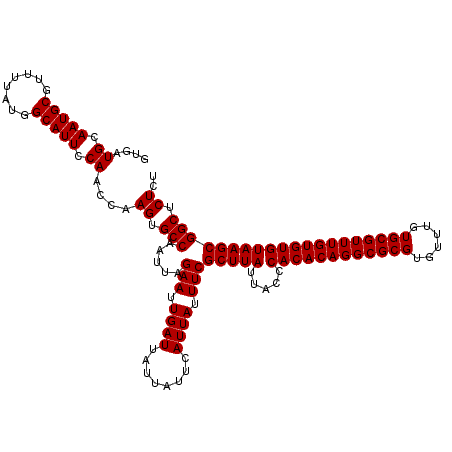
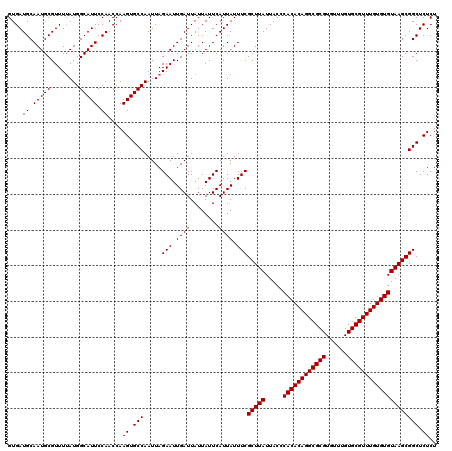
| Location | 22,362,763 – 22,362,883 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22362763 120 - 27905053 AGAGAGCCGCUUACACACAAACGCACAAACACGCGCCUGUGUGGGUAAUAAGCGAAAUAAUGAAUAAUAAUCAAUUCUAAUUGGCACUUGGUUGGAAUGCCAUAAAACGCAUUGCAUCAC .(((.((((((((((((((..(((........)))..)))))).....)))))........((((........)))).....))).)))(((.(.(((((........))))).)))).. ( -30.40) >DroSim_CAF1 74459 120 - 1 AGAGAGCCGCUUACACACAAACGCACGAACACGCGCCUGUGUGGGUAAUAAGCGAAAUAAUGAAUAAUAAUCAAUUCUAAUUGGCACUUGGUUGGAAUGCCAUAAAACGCAUUGCAUCAC .(((.((((((((((((((..(((........)))..)))))).....)))))........((((........)))).....))).)))(((.(.(((((........))))).)))).. ( -30.40) >DroEre_CAF1 73265 120 - 1 AGAGAGCCGCUUACACACAAACGCACAAACACGCGCCUGUGUGGGUAAUAAGCGAAAUAAUGAAUAAUAAUCAAUUCUAAUUGGCACUUGGUUGGAAUGCCAUUAAACGCAUUGCAUCAC .(((.((((((((((((((..(((........)))..)))))).....)))))........((((........)))).....))).)))(((.(.(((((........))))).)))).. ( -30.40) >DroYak_CAF1 72806 120 - 1 AGAGAGCCGCUUACACACAAACGCACAAACACGCGCCUGUGUGGGUAAUAAGCGAAAUAAUGAAUAAUAAUCAAUUCUAAUUGGCACUUGGUUGGAAUGCCAUAAAACGCAUUGCAUCAC .(((.((((((((((((((..(((........)))..)))))).....)))))........((((........)))).....))).)))(((.(.(((((........))))).)))).. ( -30.40) >consensus AGAGAGCCGCUUACACACAAACGCACAAACACGCGCCUGUGUGGGUAAUAAGCGAAAUAAUGAAUAAUAAUCAAUUCUAAUUGGCACUUGGUUGGAAUGCCAUAAAACGCAUUGCAUCAC .(((.((((((((((((((..(((........)))..)))))).....)))))........((((........)))).....))).)))(((.(.(((((........))))).)))).. (-30.40 = -30.40 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:31 2006