
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,359,156 – 22,359,254 |
| Length | 98 |
| Max. P | 0.995051 |

| Location | 22,359,156 – 22,359,254 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
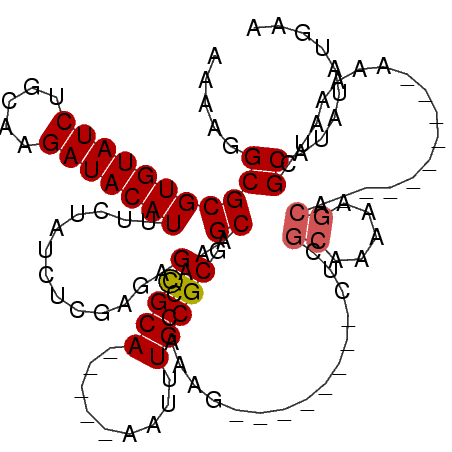
>3R_DroMel_CAF1 22359156 98 + 27905053 AAAAGGCGCGUGUAUCUGCAAGAUACAUUUCUAUCUCAAGAGCGCA-----AAAUUGCGCGGCAAACCAAAG--------CUCGCAAAAAGCA---------AAAAAUCGCAUAUAUGAA .....(((((((((((.....))))))).(((......))))))).-----...(((((.(((........)--------)))))))...((.---------.......))......... ( -24.50) >DroSim_CAF1 70858 98 + 1 AAAAGGCGCGUGUAUCUGCAAGAUACAUUUCUAUCUCGAGAGCGCA-----AAUUUGCGCGGCAAACCAAAG--------CUCGCAAAAAGCG---------AAAAAUCGCAUAUAUGAA .....(((((((((((.....))))))).(((......))))))).-----..((((((.(((........)--------))))))))..(((---------(....))))......... ( -29.10) >DroEre_CAF1 69350 95 + 1 AAAAGGCGCGUGUAUCUUCAAGAUACAUUUCUAUCUCGAGAGCGCA-----AAUUUGCGCGGCAAACCGAAG--------CUCGCC---AGCA---------AAAAAUCGCAUAUAUGAA ....(((..(((((((.....))))))).........(((.(((((-----....)))))((....))....--------))))))---.((.---------.......))......... ( -25.60) >DroWil_CAF1 75030 118 + 1 -AAAGGCGCGUGUAUCUAGAAGAUACAUUUCUAGCUU-AGAGUGCACGACGAAUUUGCGCGGCAAGAAAAAAAAAAUAAAGACGAAAAAAGAACUAAAUAAAUAAAGUCGCAUAAAUGAA -....((((.((((((.....))))))(((((.(((.-...(((((.........)))))))).))))).....................................).)))......... ( -20.30) >DroYak_CAF1 68858 95 + 1 AAAAGGCGCGUGUAUCUCGAAGAUACAUUUCUAUCUCGAGAGCGCA-----AAUUUGCGCGGCAAACCGAAG--------CUCGCC---AGCA---------AAAAAUCGCAUAUAUGAA ....((((.((...((((((.((((......))))))))))(((((-----....)))))((....))...)--------).))))---.((.---------.......))......... ( -29.10) >consensus AAAAGGCGCGUGUAUCUGCAAGAUACAUUUCUAUCUCGAGAGCGCA_____AAUUUGCGCGGCAAACCAAAG________CUCGCAAAAAGCA_________AAAAAUCGCAUAUAUGAA .....(((((((((((.....))))))).............(((((.........))))).))....................((.....)).................))......... (-17.34 = -17.58 + 0.24)

| Location | 22,359,156 – 22,359,254 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -16.56 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
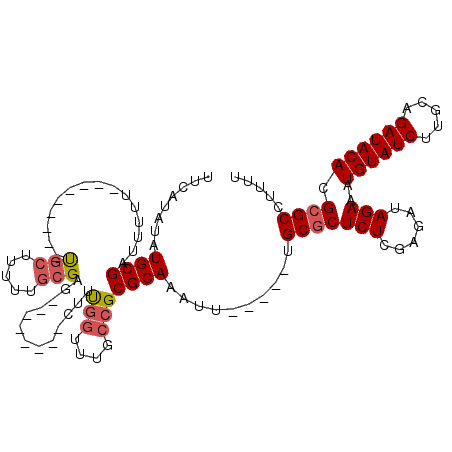
>3R_DroMel_CAF1 22359156 98 - 27905053 UUCAUAUAUGCGAUUUUU---------UGCUUUUUGCGAG--------CUUUGGUUUGCCGCGCAAUUU-----UGCGCUCUUGAGAUAGAAAUGUAUCUUGCAGAUACACGCGCCUUUU .........(((..((((---------(((((...(((((--------(....)))))).(((((....-----)))))....))).))))))((((((.....)))))))))....... ( -26.00) >DroSim_CAF1 70858 98 - 1 UUCAUAUAUGCGAUUUUU---------CGCUUUUUGCGAG--------CUUUGGUUUGCCGCGCAAAUU-----UGCGCUCUCGAGAUAGAAAUGUAUCUUGCAGAUACACGCGCCUUUU .........(((...(((---------(...(((((.(((--------(...(((((((...)))))))-----...)))).)))))..))))((((((.....)))))))))....... ( -27.30) >DroEre_CAF1 69350 95 - 1 UUCAUAUAUGCGAUUUUU---------UGCU---GGCGAG--------CUUCGGUUUGCCGCGCAAAUU-----UGCGCUCUCGAGAUAGAAAUGUAUCUUGAAGAUACACGCGCCUUUU .........(((......---------....---((((((--------(....)))))))(((((....-----)))))((((((((((......))))))).)))....)))....... ( -32.70) >DroWil_CAF1 75030 118 - 1 UUCAUUUAUGCGACUUUAUUUAUUUAGUUCUUUUUUCGUCUUUAUUUUUUUUUUCUUGCCGCGCAAAUUCGUCGUGCACUCU-AAGCUAGAAAUGUAUCUUCUAGAUACACGCGCCUUU- .........(((.......................................(((((.(((((((......).))))......-..)).)))))((((((.....)))))))))......- ( -16.20) >DroYak_CAF1 68858 95 - 1 UUCAUAUAUGCGAUUUUU---------UGCU---GGCGAG--------CUUCGGUUUGCCGCGCAAAUU-----UGCGCUCUCGAGAUAGAAAUGUAUCUUCGAGAUACACGCGCCUUUU .........(((......---------....---((((((--------(....)))))))(((((....-----)))))((((((((((......)))).))))))....)))....... ( -34.80) >consensus UUCAUAUAUGCGAUUUUU_________UGCUUUUUGCGAG________CUUUGGUUUGCCGCGCAAAUU_____UGCGCUCUCGAGAUAGAAAUGUAUCUUGCAGAUACACGCGCCUUUU ........((((...............(((.....))).............(((....)))))))..........(((((((......)))..((((((.....)))))).))))..... (-16.56 = -17.16 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:22 2006