
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,357,724 – 22,357,821 |
| Length | 97 |
| Max. P | 0.791875 |

| Location | 22,357,724 – 22,357,821 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -14.45 |
| Energy contribution | -16.13 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22357724 97 + 27905053 -------UAU-----UAUUAUA---AAUCUA-----CUGAAAACACUCAAAUUUGGUCGGAA-UAACUGCCGAGUUGGCAUCACAACUGCGAAUUUAGAGUGUGAAAU--AUGGCAAAGG -------...-----.......---...(((-----......(((((((((((((.((((..-......))))((((......))))..))))))).)))))).....--.)))...... ( -21.20) >DroSec_CAF1 65440 112 + 1 AAGCGCUUAUGUUUUUACUAUACUUUAUCUG-----CUAAGAACACUCCAAUUUGGGGGGAA-UAACUGACAAAUUGGCAUCACAACUGCGAAUUUGGAGUGUGAAAU--AUGGCAGAGG ...........................((((-----(((...(((((((((((((..((...-...))..))))((.(((.......))).)).))))))))).....--.))))))).. ( -28.60) >DroSim_CAF1 69435 105 + 1 -------UAUGUUUUUACUAUAUUUUAUCUA-----CUAAAAACACUCCAAUUUGUCCGGAA-UAACUGCCAAUUUGGCAUCACAACUACGAAUUUAGAGUGUGAAAU--AUGGCAGAGG -------.....((((.(((((((((.....-----......((((((.(((((((..((..-....((((.....))))......)))))))))..)))))))))))--)))).)))). ( -21.00) >DroEre_CAF1 67790 101 + 1 -----------UUCUUAUU-UA---AAUCUGGUAGCCUAA-AACACUCCAAUUUGGGCGGCA-AAACUGCCGAAUUGGCAUCACAACUGCGAAUUCGGAGUGUGAAAU--AUGACAGAGG -----------........-..---..((((.........-.(((((((......(((((..-...)))))(((((.(((.......))).)))))))))))).....--....)))).. ( -32.31) >DroYak_CAF1 67348 105 + 1 -----------UUUGUAUU-UA---ACUCUGGUAGCCUAAAAACACUCCAAUUUGGCUGGAAAAAACUACACAAUUGGCAUCACAACUGCGAAUUCAGAGUGUGAAAUAUAUGGCAGAGG -----------........-..---.(((((.((........(((((((((((.(..(((......)))..))))))(((.......))).......))))))........)).))))). ( -22.19) >consensus _______UAU_UUUUUAUUAUA___AAUCUG_____CUAAAAACACUCCAAUUUGGCCGGAA_UAACUGCCAAAUUGGCAUCACAACUGCGAAUUUAGAGUGUGAAAU__AUGGCAGAGG ...........................((((...........((((((((((((((.(((......)))))))))))(((.......))).......))))))...........)))).. (-14.45 = -16.13 + 1.68)

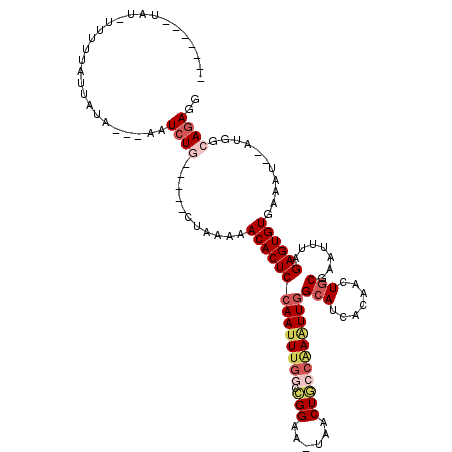
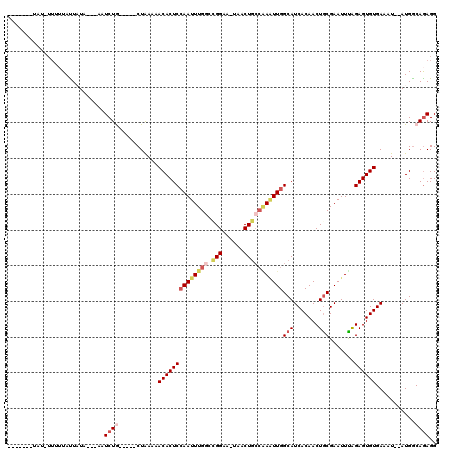
| Location | 22,357,724 – 22,357,821 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.39 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22357724 97 - 27905053 CCUUUGCCAU--AUUUCACACUCUAAAUUCGCAGUUGUGAUGCCAACUCGGCAGUUA-UUCCGACCAAAUUUGAGUGUUUUCAG-----UAGAUU---UAUAAUA-----AUA------- ..(((((...--.....((((((.(((((.(..(..((((((((.....)))).)))-)..)...).))))))))))).....)-----))))..---.......-----...------- ( -17.12) >DroSec_CAF1 65440 112 - 1 CCUCUGCCAU--AUUUCACACUCCAAAUUCGCAGUUGUGAUGCCAAUUUGUCAGUUA-UUCCCCCCAAAUUGGAGUGUUCUUAG-----CAGAUAAAGUAUAGUAAAAACAUAAGCGCUU ..(((((...--.....(((((((....((((....))))....((((((.......-.......))))))))))))).....)-----))))........................... ( -19.96) >DroSim_CAF1 69435 105 - 1 CCUCUGCCAU--AUUUCACACUCUAAAUUCGUAGUUGUGAUGCCAAAUUGGCAGUUA-UUCCGGACAAAUUGGAGUGUUUUUAG-----UAGAUAAAAUAUAGUAAAAACAUA------- ...((((((.--...(((((..(((......))).)))))........))))))...-.(((((.....)))))(((((((((.-----((.........)).))))))))).------- ( -23.80) >DroEre_CAF1 67790 101 - 1 CCUCUGUCAU--AUUUCACACUCCGAAUUCGCAGUUGUGAUGCCAAUUCGGCAGUUU-UGCCGCCCAAAUUGGAGUGUU-UUAGGCUACCAGAUU---UA-AAUAAGAA----------- ..((((....--.....((((((((((((.(((.......))).)))))(((.....-.))).........))))))).-.........))))..---..-........----------- ( -25.81) >DroYak_CAF1 67348 105 - 1 CCUCUGCCAUAUAUUUCACACUCUGAAUUCGCAGUUGUGAUGCCAAUUGUGUAGUUUUUUCCAGCCAAAUUGGAGUGUUUUUAGGCUACCAGAGU---UA-AAUACAAA----------- .(((((...........((((((..(...((((((((......))))))))..(((......)))....)..))))))...........))))).---..-........----------- ( -22.15) >consensus CCUCUGCCAU__AUUUCACACUCUAAAUUCGCAGUUGUGAUGCCAAUUCGGCAGUUA_UUCCGCCCAAAUUGGAGUGUUUUUAG_____CAGAUU___UAUAAUAAAAA_AUA_______ ..((((...........(((((((((..((((....))))((((.....))))................)))))))))...........))))........................... (-17.91 = -17.39 + -0.52)

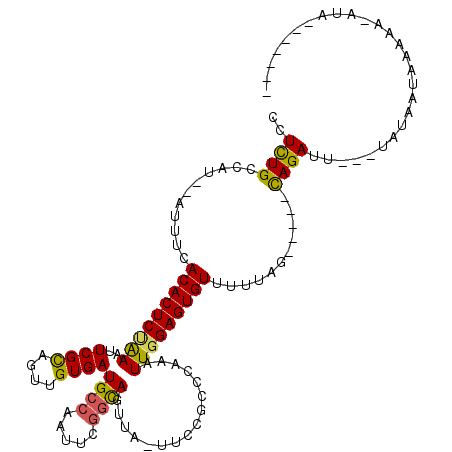
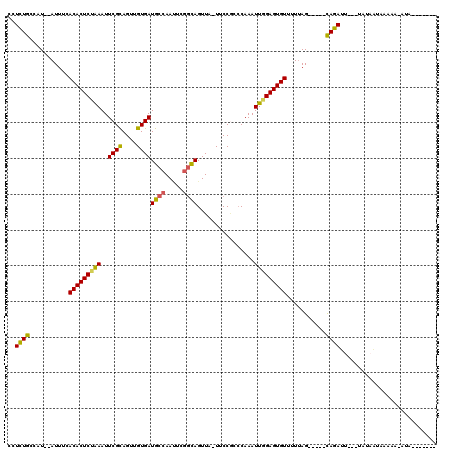
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:17 2006