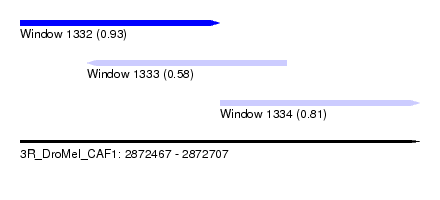
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,872,467 – 2,872,707 |
| Length | 240 |
| Max. P | 0.933506 |

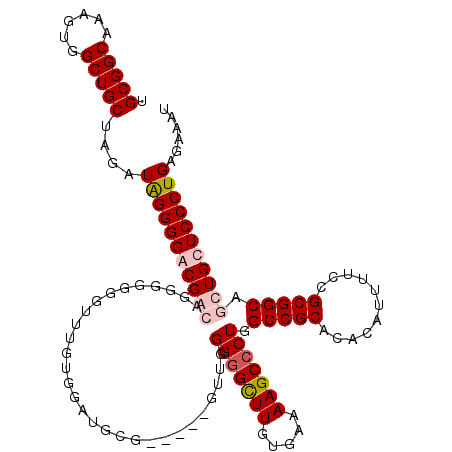
| Location | 2,872,467 – 2,872,587 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.57 |
| Mean single sequence MFE | -48.15 |
| Consensus MFE | -38.39 |
| Energy contribution | -39.70 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2872467 120 + 27905053 UGCGGCAAAGUGGCUGCUAGAUAGGGGGCGAGAGGGGGUGUUUGGGGAUGGGUGGGUGUUGGGGGCUUGUGAAAAGCCCUGGCCGCACACAUUUUCCGCGGCAGCUGCUCCCUGAGAAAU .(((((......)))))....((((((((.((....(.(((..(..((((.(((.(.((..(((.(((.....))))))..))).))).))))..).))).)..))))))))))...... ( -50.40) >DroSec_CAF1 3670 115 + 1 UGCGGCAAAGUGGCUGCUAGAUGGGGGACGGCAGAGGGGGUUUGUGGAUGCG-----GUUGGGGGCUUGUGAAAAGCCCUGGCCGCACACAUUUUCCGCGGCAGCUGCUCCCUGAGAAAU .(((((......))))).....(((((.((((.(..(.((...(((..((((-----((..(((.(((.....))))))..))))))..)))...)).)..).)))))))))........ ( -54.00) >DroSim_CAF1 4051 116 + 1 UGCGGCAAAGUGGCUGCUAGAUGGGGGACGGCAGGGGGGGUUUGUGGAUGCG----GGUUGGGGGCUUGUGAAAAGCCCUGGCCGCACACAUUUUCCGCGGCAGCUGCUCCCUGAGAAAU .(((((......))))).....(((((.((((.(.((..(..((((..((((----(.(..(((((((.....)))))))).))))))))).)..)).)....)))))))))........ ( -51.40) >DroEre_CAF1 4616 100 + 1 UGCGGCAAAGUGGCUGCUAGAUAGGGGGCGAGGGGGG----------A----------UGGGGAGUUUCUGAAAAGCCCUGGCCGCACACAUUUUCCGCGGCAGCUGCUCCCUGAGAAAU .(((((......)))))....(((((((((..((..(----------(----------(((((..((.....))..)))((......))))))..))((....)))))))))))...... ( -36.80) >consensus UGCGGCAAAGUGGCUGCUAGAUAGGGGACGACAGGGGGGGUUUGUGGAUGCG_____GUUGGGGGCUUGUGAAAAGCCCUGGCCGCACACAUUUUCCGCGGCAGCUGCUCCCUGAGAAAU .(((((......)))))....(((((((((((.............................(((((((.....))))))).(((((...........))))).)))))))))))...... (-38.39 = -39.70 + 1.31)


| Location | 2,872,507 – 2,872,627 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -29.81 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2872507 120 - 27905053 CGACUUCCUUACUAACUACGCACGAAAUCAAGUGGGGAAAAUUUCUCAGGGAGCAGCUGCCGCGGAAAAUGUGUGCGGCCAGGGCUUUUCACAAGCCCCCAACACCCACCCAUCCCCAAA ....(((((((((.................))))))))).........((((...(((((((((.....)))).)))))..((((((.....))))))..............)))).... ( -34.23) >DroSec_CAF1 3710 115 - 1 CGACUUCCUUACUAACUACGCACGAAAUCAAGUGGGGAAAAUUUCUCAGGGAGCAGCUGCCGCGGAAAAUGUGUGCGGCCAGGGCUUUUCACAAGCCCCCAAC-----CGCAUCCACAAA ....(((((((((.................)))))))))..........(((((.(((((((((.....)))).)))))..((((((.....)))))).....-----.)).)))..... ( -34.73) >DroSim_CAF1 4091 116 - 1 CGACUUCCUUACUAACUACGCACGAAAUCAAGUGGGGAAAAUUUCUCAGGGAGCAGCUGCCGCGGAAAAUGUGUGCGGCCAGGGCUUUUCACAAGCCCCCAACC----CGCAUCCACAAA ....(((((((((.................)))))))))..........(((((.(((((((((.....)))).)))))..((((((.....))))))......----.)).)))..... ( -34.73) >DroEre_CAF1 4653 103 - 1 CGACUUCCUUACUAACUACGCACGAAAUCAAGUGGGGAAAAUUUCUCAGGGAGCAGCUGCCGCGGAAAAUGUGUGCGGCCAGGGCUUUUCAGAAACUCCCCA----------U------- ...............................(((((((...(((((..((((((..((((((((.(.....).))))).))).)))))).))))).))))))----------)------- ( -30.50) >consensus CGACUUCCUUACUAACUACGCACGAAAUCAAGUGGGGAAAAUUUCUCAGGGAGCAGCUGCCGCGGAAAAUGUGUGCGGCCAGGGCUUUUCACAAGCCCCCAAC_____CGCAUCCACAAA .(.((((((...........(((........)))((((.....))))))))))).(((((((((.....)))).)))))..((((((.....))))))...................... (-29.81 = -29.88 + 0.06)

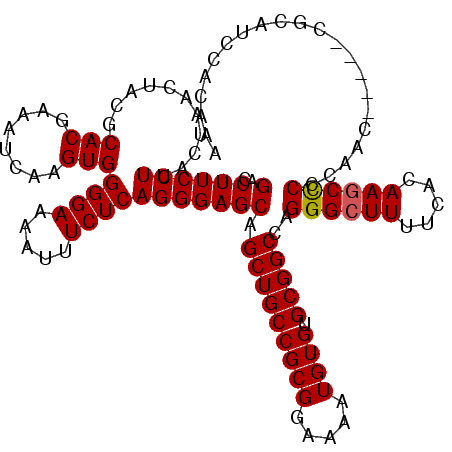

| Location | 2,872,587 – 2,872,707 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -28.85 |
| Energy contribution | -30.85 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2872587 120 + 27905053 UUUCCCCACUUGAUUUCGUGCGUAGUUAGUAAGGAAGUCGCGUUCAUUCGUUCGCAGCUCUAACGAGCCCCAGCGAAUAUUUAUGAUCAGCACGGGCUCAUGGGCCUCCUCCUAACCACC ......(((........))).((.(((((..((((.((.((..((((..((((((.((((....))))....))))))....))))...)))).((((....)))))))).))))).)). ( -35.30) >DroSec_CAF1 3785 119 + 1 UUUCCCCACUUGAUUUCGUGCGUAGUUAGUAAGGAAGUCGCGUUCAUUCGUUCGCAGCUCUAACGAGCCCCAGCGAAUAUUUAUGAUCAGCAUGGGCUCACCG-CCCACUCCUCUCCACC ...........((((((.(((.......)))..))))))((..((((..((((((.((((....))))....))))))....))))...)).(((((.....)-))))............ ( -34.80) >DroSim_CAF1 4167 116 + 1 UUUCCCCACUUGAUUUCGUGCGUAGUUAGUAAGGAAGUCGCGUUCAUUCGUUCGCAGCUCUAACGAGCCCCAGCGAAUAUUUAUGAUCAGCAUGGGCUCAC----CCACUCCUCACCACC ....((((...((((((.(((.......)))..))))))((..((((..((((((.((((....))))....))))))....))))...)).)))).....----............... ( -31.70) >DroEre_CAF1 4716 117 + 1 UUUCCCCACUUGAUUUCGUGCGUAGUUAGUAAGGAAGUCGCGUUCAUUCGUUCGCAGCUCUAACGAGCCCCAGCGAAUAUUUAUGAUCAGCCUGGGCUCACCG-CCC--CCUUUACCACC ...........((((((.(((.......)))..))))))((..((((..((((((.((((....))))....))))))....))))...))..((((.....)-)))--........... ( -32.30) >consensus UUUCCCCACUUGAUUUCGUGCGUAGUUAGUAAGGAAGUCGCGUUCAUUCGUUCGCAGCUCUAACGAGCCCCAGCGAAUAUUUAUGAUCAGCAUGGGCUCACCG_CCCACUCCUCACCACC ....(((....((((((.(((.......)))..))))))((..((((..((((((.((((....))))....))))))....))))...))..)))........................ (-28.85 = -30.85 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:24 2006