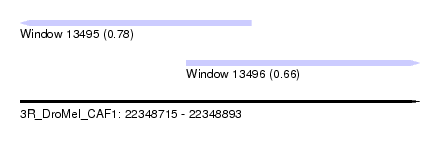
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,348,715 – 22,348,893 |
| Length | 178 |
| Max. P | 0.775221 |

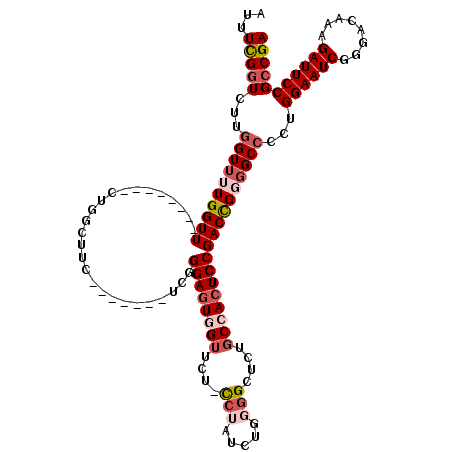
| Location | 22,348,715 – 22,348,818 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.29 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -21.89 |
| Energy contribution | -21.33 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
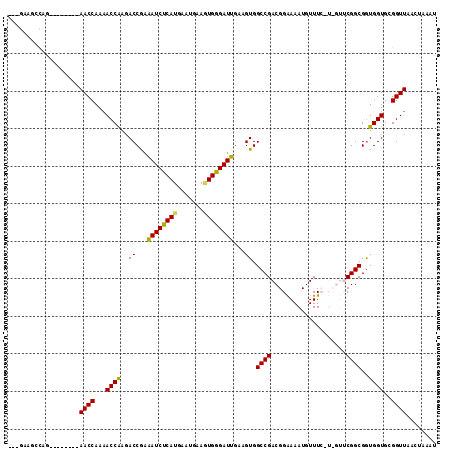
>3R_DroMel_CAF1 22348715 103 - 27905053 ---GGAGCCAG--------AACCAAAACCAAGACCAAAAUCUCAUGAAUGAAGUGGGAUUGAAGUGGCCGACGGAAAAUGUUUCAUCGUUCGGCGGUGGUGCGGUUAACUAAAU ---..((((..--------.(((...(((...((...((((((((.......))))))))...)).((((((((((.....)))..)).)))))))))))..))))........ ( -26.60) >DroSec_CAF1 56487 103 - 1 ---GAAGCCAG--------AACCAAAACCAAGACCGAAAUCUCAUGAAUGAAAUGGGAUUGAAGUGGCCGACGGAAAAUGUUUCGUUGUUCGGCGGUGGUGCGGUUAACUAAAU ---........--------((((...((((.(.((((((((((((.......))))))))........((((((((....)))))))).)))))..))))..))))........ ( -28.90) >DroSim_CAF1 55629 103 - 1 ---GAAGCCAG--------AACCAAAACCGAGACCGAAAUCUCAUGAAUGAAGUGGGAUUGAAGUGGCCGACGGAAAAUGUUUCGUCGUUCGGCGGUGGUGCGGUUAACUAAAU ---..((((..--------.((((...(((((.(((.((((((((.......))))))))....))).((((((((....)))))))))))))...))))..))))........ ( -33.80) >DroEre_CAF1 58691 103 - 1 ---GAAUCUAA-AUCCAACAACCAACACCAAAACCGAGAUCCCAUGAAUGAAUUGGGAUUGAAGUGGCCGACGGGAAAUGUUU-------CGGCGAUGGUGCGGUUAACUAAAU ---........-.......((((..(((((..((...(((((((.........)))))))...)).(((((...........)-------))))..))))).))))........ ( -26.40) >DroYak_CAF1 58159 105 - 1 CGUGAACCUAAAAUCCA--AACCAAAACCGAAACCAAAAUCCCAUGAAUGAAGUGGGAUUGAAGUGGCCGACGGAAAAUGUUU-------CGGCGGUGGUGCGGUUAACUAAAU .................--......(((((..(((..((((((((.......))))))))......((((.(((((....)))-------)).))))))).)))))........ ( -28.50) >consensus ___GAAGCCAG________AACCAAAACCAAGACCGAAAUCUCAUGAAUGAAGUGGGAUUGAAGUGGCCGACGGAAAAUGUUUC_U_GUUCGGCGGUGGUGCGGUUAACUAAAU ...................((((...((((..((...((((((((.......))))))))...)).((((....................))))..))))..))))........ (-21.89 = -21.33 + -0.56)

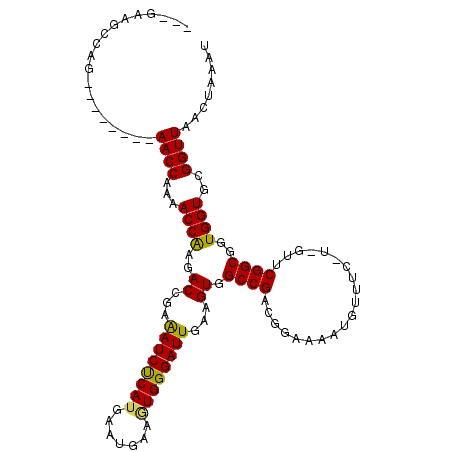
| Location | 22,348,789 – 22,348,893 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -27.56 |
| Energy contribution | -29.24 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22348789 104 + 27905053 AUUUUGGUCUUGGUUUUGGUU--------CUGGCUCC-------UCGGGAGUGGUUCU-CCUAUCUGGGGCUCUGCCACUCCGACCGGGGCCCCUGGAAUCGGGACAAAGAUUCCGACGA ...((((((((.(((((((((--------((((((((-------((((.((((((.((-((.....))))....))))))))))..))))))...))))))))))).))))..))))... ( -46.90) >DroSec_CAF1 56561 104 + 1 AUUUCGGUCUUGGUUUUGGUU--------CUGGCUUC-------UCGGGAGUGGUUCU-UCUAUCUGGGGCUCUGCCACUCCGACCGGGGCCCCUGGAAUCGGGACAAAGAUUCCGCCGA ...((((((((.(((((((((--------((((((((-------((((.((((((.((-((.....))))....))))))))))..))))))...))))))))))).))).....))))) ( -41.80) >DroSim_CAF1 55703 105 + 1 AUUUCGGUCUCGGUUUUGGUU--------CUGGCUUC-------UCGGGAGUGGUUCUCUGGGUCUGGGGCUCUGCCACUCCGACCGGGGCCCCUGGAAUCGGGACAAAGAUUCCGCCGA ...(((((...((....((((--------((((....-------((((.((((((.....((((.....)))).)))))))))))))))))))).((((((........))))))))))) ( -43.50) >DroEre_CAF1 58758 109 + 1 AUCUCGGUUUUGGUGUUGGUUGUUGGAU-UUAGAUUC-------UCGGGAGUGGUUCU-CCUAUCUGCGGUCUG-CGAAUCCGACUG-GGCCCCUGGAAUCGGGACAAAGAUUCCGCCGA ...(((((...((.(((.(..(((((((-((((((..-------..(((((.....))-)))))))((.....)-))))))))))).-))).)).((((((........))))))))))) ( -35.60) >DroYak_CAF1 58226 116 + 1 AUUUUGGUUUCGGUUUUGGUU--UGGAUUUUAGGUUCACGGGAGUCGGGAGUGGUUCU-CCUAUCUGGGGUCUGGCGACUCCGACUG-GGCACCUGGAAUCGGGACAAAGAUUCCGCCGA ...((((...(((..((.(((--(.((((((((((((.(((.((..(((((.....))-)))..))((((((....))))))..)))-)).)))))))))).))))...))..))))))) ( -43.70) >consensus AUUUCGGUCUUGGUUUUGGUU________CUGGCUUC_______UCGGGAGUGGUUCU_CCUAUCUGGGGCUCUGCCACUCCGACCGGGGCCCCUGGAAUCGGGACAAAGAUUCCGCCGA ...(((((...((((((((((..........................((((((((....(((.....)))....))))))))))))))))))...((((((........))))))))))) (-27.56 = -29.24 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:04 2006